AWS News Blog
New AWS Public Data Set – 3000 Rice Genome
|
|
My colleague Angel Pizarro wrote the guest post below to tell you about an amazing new AWS Public Data Set!
— Jeff;
You can now access the genome sequence data of 3,024 rice varieties that have been aligned and analyzed against five different reference genomes as an AWS Public Data Set. The data contains over 30 million genetic variations that span across all known and predicted rice genes, as well as potential regulatory regions surrounding these genes. Through analysis of this data, researchers can potentially identify genes associated with important agronomic traits such as crop yield, climate stress tolerance, and disease resistance. Together, they represent an unprecedented resource for advancing rice science and breeding technology.
Rice is a staple food source for half the world’s population, and accounts for over 20% of all calories per capita. In order to keep up with global population increases, we must find some way to increase rice crop yields by 25% by 2030. The current rate of increasing rice yield by traditional breeding is insufficient, especially when taking into account observed trends in climate change and pollution. In order to meet the world’s projected demand for a stable food supply, modern methods of breeding that take into account the underlying genetic information must be adopted by the community at large.
The 3,000 Rice Genome sequencing project is an international effort to sequence the genomes of 3,024 rice varieties from 89 countries. The collaborating centers involved are the Chinese Academy of Agricultural Sciences, BGI Shenzhen, and the International Rice Research Institute (IRRI). The consortium partnered with DNAnexus to analyze the sequence data of the 3,024 different rice varieties against five published draft genome builds of the rice genome. Partnering with DNAnexus allowed them to take advantage of the scalable computing capability at AWS to process all of the source genomic data across 37,000 compute cores working together in just two days — more than 200 times faster than would have been possible on local computing infrastructure. In addition, the data are accessible via DNAnexus for further analysis. For more details on accessing the data within DNAnexus, refer to the project documentation.
More in-depth analyses of this dataset could lead to inferences about higher yield and stress tolerance to pests, diseases, and climate change. You can learn more about the data and how to access it on the 3000 Rice Genome Public Data Set page.
Working with the Genomic Data Set on AWS
Because the data are hosted on S3 and accessible over common HTTP protocols, researchers have already done some amazing integrations within pre-existing tools. I’ve included some initial examples here and we’ll work with IRRI to share more examples as they emerge.
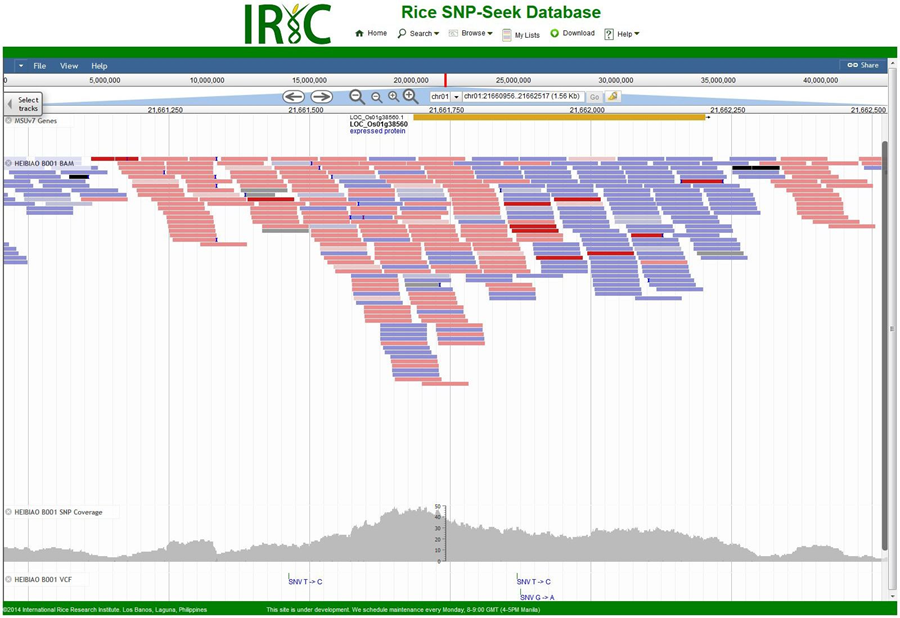
Visualizing the data using SNP-Seek
The International Rice Informatics Consortium (IRIC) has made the data available for querying and visualization through their SNP-Seek portal. User are now able to query across all of the strains and narrow down regions of interest that show diversity across multiple genome references, integrated with the rice research community’s genomic annotation data:
Open Source Tools
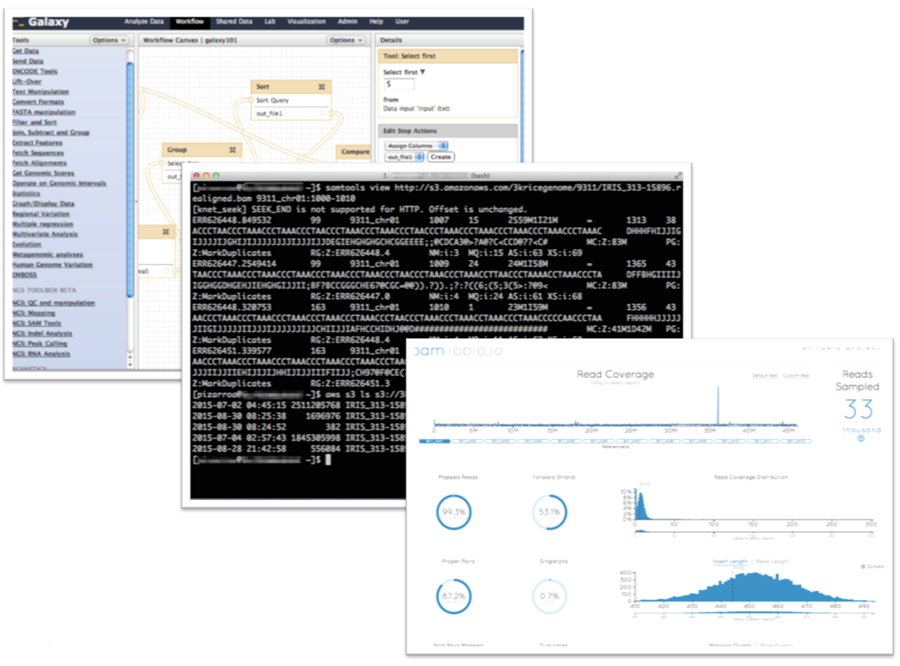
In addition to the rich set of AWS partner offerings for life sciences, the full genomics open source ecosystem is available for use with the data. From command line applications such as samtools to rich user interfaces such as Galaxy or iobio, researchers can get started right away to analyze the data.
What’s Next?
The challenge for the research community is now to comprehensively and systematically mine this dataset to link genotypic variation to functional variation with the ultimate goal of creating new and sustainable rice varieties. Combining these efforts with other studies such as careful trait phenotyping in controlled and wild environments, as well as environmental studies based on satellite imagery like the Landsat data you can already access on AWS, can help is to keep up with the demands of the future world’s population growth.
Visit the 3000 Rice Genome Public Data Set page to access the data and sign up for project updates.
— Angel Pizarro, Technical Business Development Manager, AWS Scientific Computing
