AWS Machine Learning Blog
Amazon SageMaker supports kNN classification and regression
We’re excited to announce that starting today Amazon SageMaker supports a built-in k-Nearest-Neighbor (kNN) algorithm for solving classification and regression problems. kNN is a simple, interpretable, and surprisingly strong model for multi-class classification, ranking, and regression.
Introduction to kNN
The idea behind kNN is that similar data points should have the same class, at least most of the time. This method is very intuitive and has proven itself in many domains, including recommendation systems, anomaly detection, and image/text classification.
For example, consider a case where you want to classify an image as one of 2000 possible types such as “person,” “animal,” “outdoors,” “ocean,” “sunset,” and so on. Given a proper distance function between images, the classification of an unlabeled image can be determined by the labels assigned to its nearest neighbors, meaning the labeled images that are closest to it, according to the distance function.
Another use of kNN, though less common, is for regression problems. Here, the objective is not to determine the class of a query but rather to predict a continuous number (e.g., the salary of a person, the cost of an experiment, etc.). The prediction of a query is set as a function of the labels of its neighbors. Typically it is set as the average, and at times, as the median or geometric average as methods to deal with outliers.
When should I use kNN?
We’ll walk you through a few pros and cons you should consider when you’re thinking about using kNN classification and regression.
Pros
Prediction quality: A kNN classifier is able to recover unstructured partitions of the space, as opposed to, say, a linear classifier that requires a linear separation between the classes. It can also adapt to different densities in the space, making it more robust than methods such as support vector machine (SVM) classification with radial basis function (RBF) kernel. The following two examples of 2D data illustrate different partitions of the space imposed by labeled data and the prediction of a kNN model on that space.
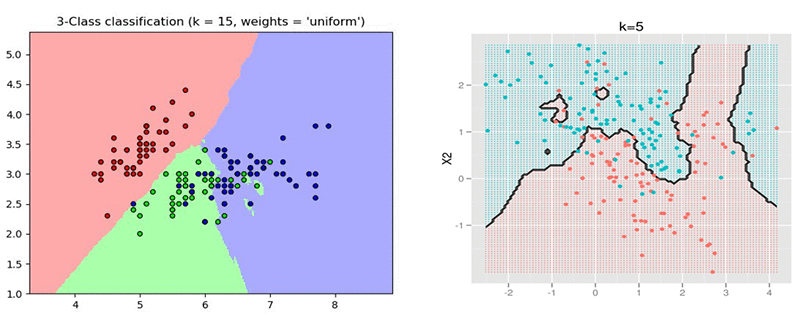
Short cycles: Another advantage of kNN is that there is little to no training involved. This means that iterating over different possible metrics / modifications of the input dataset is potentially faster when compared to a classifier that requires a heavy training procedure, such as deep networks, or even SVM or linear functions.
Many output classes: kNN can seamlessly handle a very large number of classes. For comparison, a linear model or a deep neural network with a cross-entropy loss must explicitly compute a score for each possible class, and choose the best one. Imagine, for example, a task of recognizing flower types by their image or by a set of their features, for which we have 100K labeled examples of features and 5K examples of flower types. Any model that explicitly learns 5K sets of parameters to identify each flower type could be easily prone to overfit, meaning it could produce wrong answers.
Interpretability: The prediction of a kNN model is based on similarity to existing objects. As a result, the question “why was my example given class X?” is answered by “because similar items are labeled with X.” For example, consider a model assessing the risk involved with a loan. In the example, a customer was judged to be high risk because 8 out of 10 customers who have been previously evaluated and were most similar to them in terms of X, Y, and Z, were found to be high risk. By observing the nearest neighbors, you can see objects similar to the example that are labeled as X, and you can decide whether the prediction makes sense.
Cons
Costly inference: The major disadvantage of kNN is its costly inference. To infer the label of an input query, we must find the data points closest to it. A naive solution would keep all data points in memory, and, given a query, compute the distance between it and all data points. For concrete quantities, if the training set contained n data points of d dimensions, this process requires O(nd) arithmetic operations per query and O(nd) memory. For large datasets this can be costly. This cost can be reduced, and we elaborate on that later on, but it is typically still large compared to alternative algorithms such as linear learner or decision trees.
Reducing inference cost
As mentioned earlier, inference can be quite costly with kNN. The following are common approaches used to lighten that cost, all of which are supported in Amazon SageMaker kNN.
Subsampling: A very simple, yet often very effective way of reducing the inference cost is to subsample the data. Just as in other learning settings, we might have more data points than we actually need. For example, we might have an available dataset of, say, 10M data points, but we can do a good enough job with 100K data points.
Dimension reduction: For some distances, such as L2 and cosine distance, it’s possible to reduce the dimension of the data points while maintaining the distance between a query point and any data point that is approximately the same. The quality of the approximation depends only on the dimension of the output. Small dimension means a crude approximation, yet it is often the case that we can obtain a good enough approximation for the distances while reducing the dimension. The main disadvantage of this method is that the output is dense, so for highly sparse data or data that had a low dimension to begin with, this might not be the best technique.
Quantization: Another method of reducing space and computational overhead is to work with a lower precision. You could move to float16, fixed point precision (integers), or even binary values. A similar method is called product-quantization (https://lear.inrialpes.fr/pubs/2011/JDS11/jegou_searching_with_quantization.pdf), which does not quantize individual numbers but small groups of numbers. This technique allows you to save space and computation even further than single bit quantization. The major disadvantage of this method is that its guarantees are data dependent and the hyperparameters have to be tuned. That being said, these techniques are very commonly used and are proven empirically.
Avoiding Regions Quickly: The methods we have discussed help reduce the time to compute the distance from each data point to a query. This means that the runtime per query becomes O(np) instead of O(nd), for some p << d. If the number of data points n is very large, these approaches won’t help and something else must be done. A common approach for disqualifying data points quickly is through clustering. If the center of a cluster is far away from the query we can disqualify the entire cluster without looking into all of its data points. For this technique, the data must be pre-processed to obtain m << n centers, typically with k-means clustering. Then, when a query arrives we compute its distance to all of the centers, and disregard all points that belong to clusters with centers far away from the query. This process is illustrated in the following figure. Here, the query is the green point, the data points are blue, and the cluster centers are red. The left cluster is eliminated with a single distance computation.

Amazon SageMaker kNN
Amazon SageMaker kNN has a training and an inference phase. In the training phase we collect data points to a single machine and construct a search index. A search index is the object used to obtain the (approximate) nearest neighbors of a query point. In the inference phase, we produce predictions for queries, based on their nearest neighbors, provided by the pre-built index.
Training
The training process can be done over one or more machines. During training each machine processes a shard of the data. The machines collectively obtain a large sample of the data points to be indexed. We note that by setting the sample_size hyperparameter to a value greater than or equal to the number of data points, all data points will be considered. Before communicating the sample, the machines can reduce the space of each point by invoking a dimension reduction technique. This can be set using the hyperparameters dimension_reduction_type and dimension_reduction_target, which determine the technique and the target dimension.
After the sample of data points is collected, we process it to build an index. We currently support a FAISS index, and surface its hyperparameters. FAISS is an open-source approximate nearest neighbor search tool optimized for L2 and cosine distance that supports both CPU and GPU hardware. The index types we currently support are “Flat,” “IVFFlat,” and “IVFPQ.” The documentation mentions the different hyperparameters related to each index type.
Inference
After collecting the data points and pre-processing the index as explained earlier, our model is ready for inference. The inference can either be done on CPU or GPU machines. As a rule of thumb, if you require a relatively small amount of items to be queried, meaning latency may be an issue but throughput is not, then a CPU machine is advisable. On the other hand, if high throughput is required due to many queries, it may be preferable to use a GPU machine. Inference on a GPU machine may also be advised if the index is very large. For example if your data requires 1M data points and a brute-force (“Flat”) index on a 1K dimensional data, GPU inference will likely have better latency, especially if given a batch of queries. In what follows we provide a notebook example that measures the inference run-time for several hardware types, including GPU and CPU.
The output provided for a query is not the set of nearest neighbors but rather a prediction. In a classification problem it is the most likely class. In a regression problem we return the regressed value. To allow custom predictions and exploratory analysis, we implemented a verbose inference mode that also returns the labels and distances of the nearest neighbors. In the detailed example below we show how the verbose mode is used to determine the best value for k, the number of neighbors determining the prediction.
Notable knobs
Sample size: A common scenario is that for a data set of 10M points, the best value of k is in the order of, say, 5000. In such cases, a subsample of the data containing 100K points, along with k=50 might actually give almost the same accuracy, yet the inference time is over an order of magnitude faster. As a rule of thumb, if you are considering a large value for k, odds are that sampling would cut the cost of inference while maintaining a similar accuracy.
Dimension reduction: If the data contains input vectors of dimension over 10K, it may be advisable to use a dimension reduction technique for pre-processing. This technique can reduce the dimension of the data to the order of 1000 to 5000 with negligible affect to the output quality.
Hands-on example: Classification with kNN
Let’s review an example use case for kNN. We’ll download the covtype dataset containing data for a multi-class problem. We’ll train a kNN model and apply it on a test set. To run the example yourself, take a look at the Jupyter notebook. The notebook contains the example that follows along with a deeper dive into the different hyperparameters of kNN.
Dataset
We’re about to work with the UCI Machine Learning Repository Covertype dataset (covtype) (copyright Jock A. Blackard and Colorado State University). It’s a labeled dataset where each entry describes a geographic area, and the label is a type of forest cover. There are seven possible labels, and we aim to solve the multi-class classification problem using kNN.
First, we’ll download the dataset and move it to a temporary folder.
Pre-processing the data
Now that we have the raw data, let’s process it. First load the data into numpy arrays, and randomly split it into train and test using a 90/10 split.
Upload to Amazon S3
Typically the dataset will be large and located in Amazon S3, so let’s write the data to Amazon S3 in recordio-protobuf format. First create an IO buffer that wraps the data, and then upload it to Amazon S3. Notice that the choice of Amazon S3 bucket and prefix should change for different users and different datasets.
It is also possible to provide test data. This way we can get an evaluation of the performance of the model from the training logs. To use this capability let’s upload the test data to Amazon S3 as well.
Training
We take a moment to explain at a high level, how Machine Learning training and prediction works in SageMaker. First, we need to train a model. This is a process that given a labeled dataset and hyperparameters guiding the training process, outputs a model. Once the training is done, we set up an endpoint. An endpoint is a web service that, given a request containing an unlabeled data point, or mini-batch of data points, returns a prediction or list of predictions.
In Amazon SageMaker the training is done using an object called an estimator. When setting up the estimator we specify the location (in Amazon S3) of the training data, the path (again in Amazon S3) to the output directory where the model will be serialized, generic hyperparameters such as the machine type to use during the training process, and kNN-specific hyperparameters such as the index type, etc. After the estimator is initialized, we can call its fit method to do the actual training.
Now that we are ready for training, we begin with a convenience function that starts a training job.
Next, we run the actual training job. For now, we’ll use default hyperparameters.
Notice that we mentioned a test set in the training job. When a test set is provided, the training job doesn’t just produce a model, but it also applies it to the test set and reports the accuracy. In the logs you can view the accuracy of the model on the test set. You can see the full logs in the notebook.
Setting up the endpoint
Now that we have a trained model, we are ready to run inference. The knn_estimator object contains all the information we need for hosting the model. In the following code we provide a convenience function that, given an estimator, sets up and endpoint that hosts the model. Other than the estimator object, we provide it with a name (string) for the estimator, and an instance_type. The instance_type is the machine type that will host the model. It is not restricted in any way by the hyperparameter settings of the training job.
Inference
Now that we have our predictor, let’s use it on our test dataset. The following code runs on the test dataset, computes the accuracy and the average latency. It splits up the data into 100 batches, each of size roughly 500. Then, each batch is given to the inference service to obtain predictions. After we have all predictions, we compute their accuracy given the true labels of the test set.
Deleting the endpoint
We’re now done with the example except a final clean-up act. By setting up the endpoint you started a virtual machine in the cloud, and as long as it’s not deleted the virtual machine is still running, and you are paying for it. After the endpoint is no longer necessary, you can delete it. The following code does exactly that.
Conclusion
We’ve seen how to train and host an inference endpoint for kNN. With absolutely zero tuning we obtain an accuracy of 92.2% on the covtype dataset. As a point of reference for grasping the prediction power of the kNN model, a linear model will achieve roughly 72.8% accuracy. There are several advanced issues that we did not discuss. To take a deep dive into issues such as runtime/latency and tuning the model while taking into account its accuracy and runtime efficiency, see (https://github.com/awslabs/amazon-sagemaker-examples/blob/master/introduction_to_amazon_algorithms/k_nearest_neighbors_covtype/k_nearest_neighbors_covtype.ipynb)
About the Authors
 Zohar Karnin is a Principal Scientist in Amazon AI. His research interests are in the area of large scale and online machine learning algorithms. He develops infinitely scalable machine learning algorithms for Amazon SageMaker.
Zohar Karnin is a Principal Scientist in Amazon AI. His research interests are in the area of large scale and online machine learning algorithms. He develops infinitely scalable machine learning algorithms for Amazon SageMaker.
 Amir Sadoughi is a Senior Software Development Engineer on the AWS AI SageMaker Algorithms team. He is passionate about technologies at the intersection of distributed systems and machine learning.
Amir Sadoughi is a Senior Software Development Engineer on the AWS AI SageMaker Algorithms team. He is passionate about technologies at the intersection of distributed systems and machine learning.