AWS for Industries
Element Biosciences offers Bases2FASTQ as a Ready2Run workflow on Amazon Omics
Blog is guest authored by Maxim Mass, Rosi Bajari, and Bryan Lajoie from Element Biosciences. To help customers easily build, deploy, and scale workloads, Amazon Omics now supports pre-built Ready2Run workflows from third-party software companies and open-source pipelines. Read more about the launch here.
Last year, Element Biosciences launched the AVITI benchtop sequencing instrument to offer improved accuracy and cost benefits for customers. Element has worked with Amazon Web Services (AWS) to provide customers with a fast, powerful, and cost-effective analysis workflow utilizing Element’s Ready2Run Bases2Fastq workflows on Amazon Omics. This integration enables customers to leverage a wide range of secondary analysis workflows available on the platform, starting directly from FASTQ files.
AVITI performs primary analysis onboard and allows customers to configure the instrument to stream base calls and quality scores directly to their Amazon S3 bucket. This eliminates the need for data transfer to an intermediate location or reliance on a managed and expensive subscription-based service. Element’s approach empowers customers to maintain end-to-end control of their data without upfront subscriptions to any specific service. Amazon Omics supports this by utilizing the compute and storage in the customer’s own account, while providing the scalability and benefits of cloud technology.
By employing Element Bioscience’s Bases2Fastq Ready2Run workflow on Amazon Omics, customers can rapidly perform demultiplexing to generate FASTQ files. These files can then serve as input for various secondary analysis Ready2Run workflows available through Amazon Omics. Ready2Run workflows consist of pre-built, standardized, and optimized pipelines from third-party software companies and open-source projects. Customers can easily execute these pipelines with just a few clicks or a single API call. The pricing structure for Ready2Run workflows is based on a per-run model, offering customers predictable and transparent pricing.
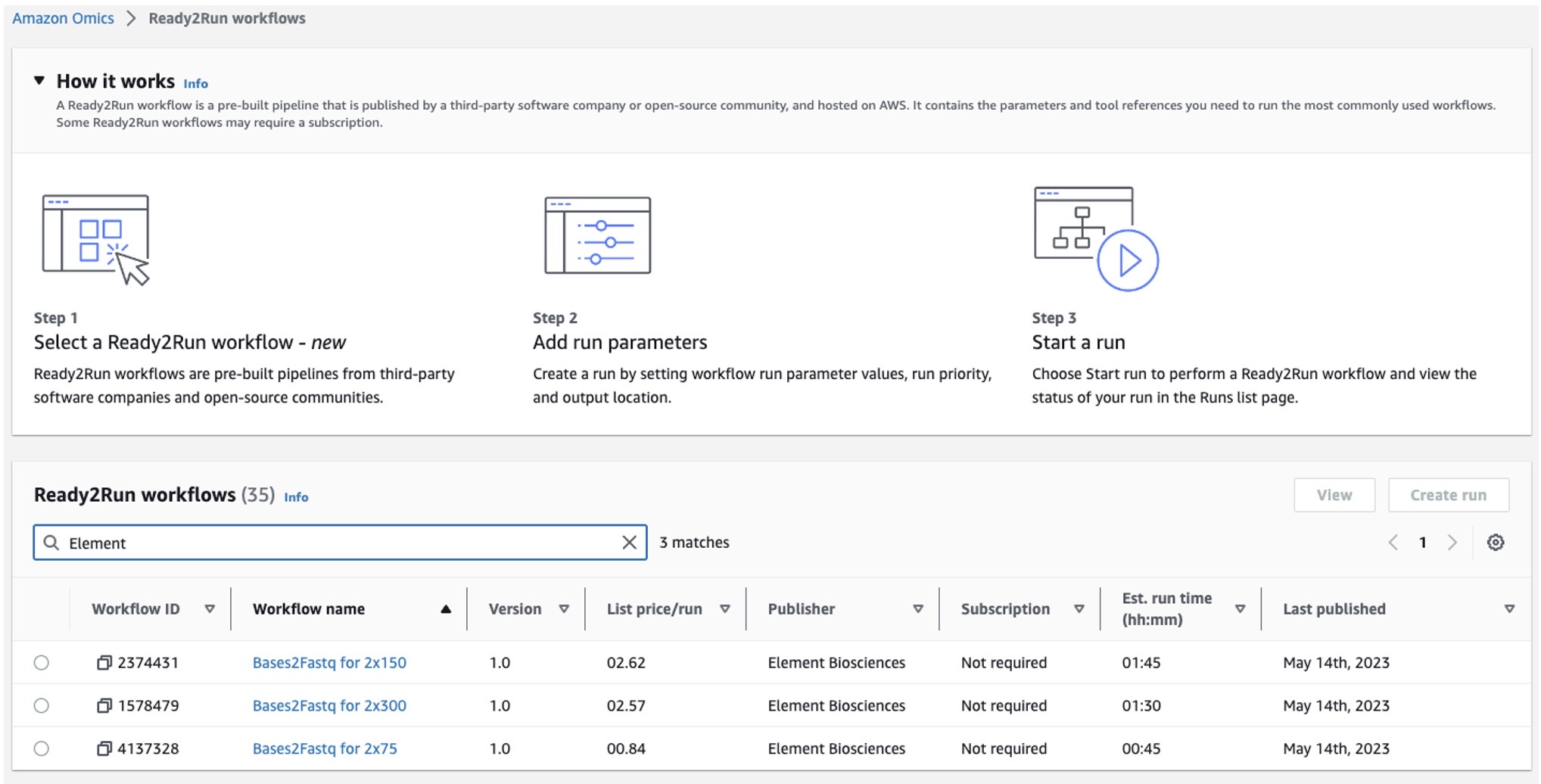 Figure 1: Element Biosciences Ready2Run workflows on Amazon Omics displaying list price per run and estimated run time
Figure 1: Element Biosciences Ready2Run workflows on Amazon Omics displaying list price per run and estimated run time
Customers can easily start a run by specifying the run directory as an input parameter for a Bases2Fastq Ready2Run workflow and the output S3 location. Once the workflow run completes, multiple files including FASTQs per sample and QC report for the run performance and quality, will be generated and stored in the customer’s specified S3 output bucket.
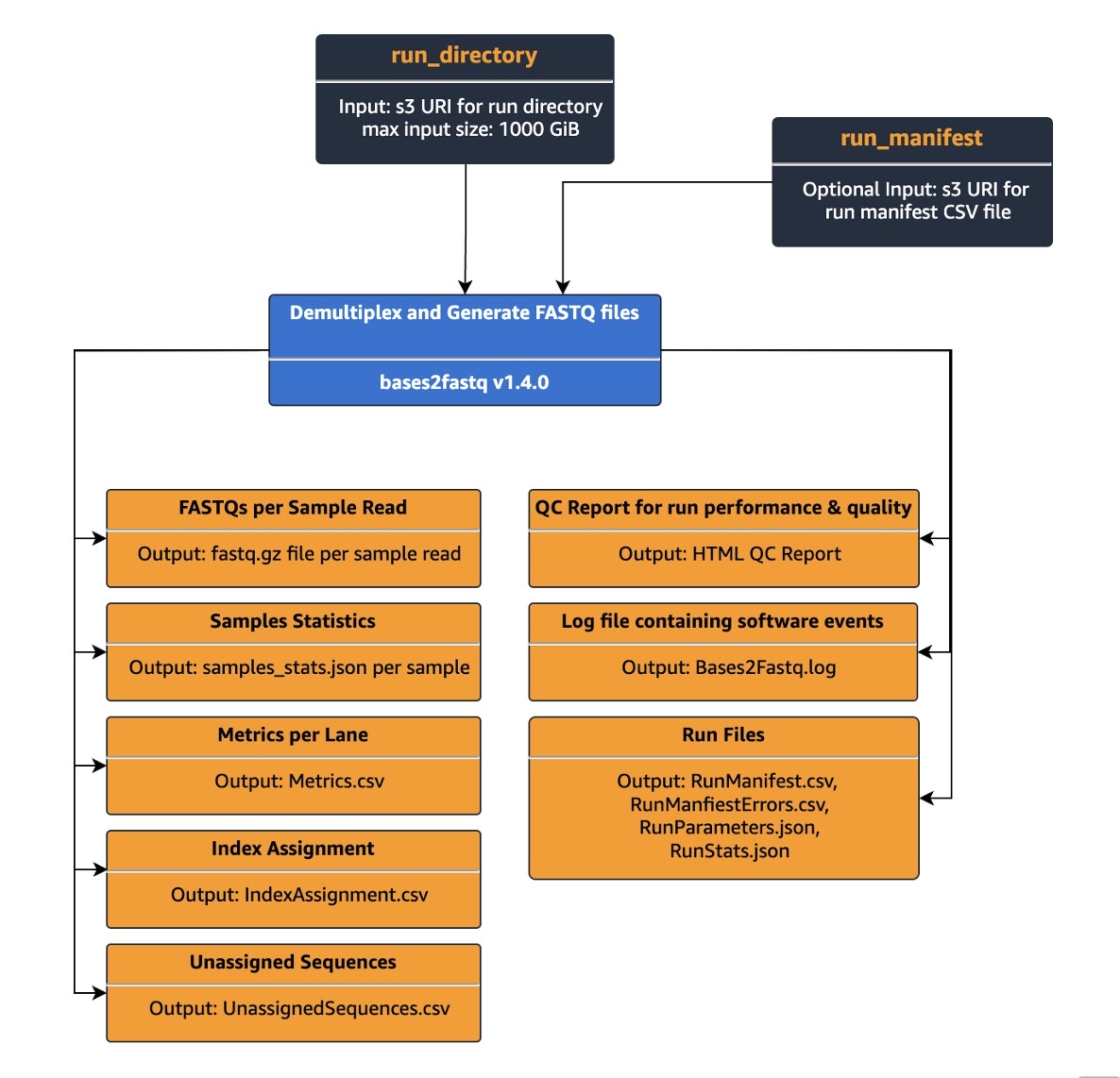
Figure 2: Bases2Fastq for 2×150 kit Ready2Run workflow diagram
Customers also have the option to utilize Element’s Bases2Fastq Ready2Run workflows through Elembio Cloud, currently in Beta. Elembio Cloud leverages Amazon Omics and other genomics analysis platforms. Using Elembio Cloud, a customer can automatically launch Bases2Fastq Ready2Run workflows using Amazon Omics in the customer’s AWS account immediately after their sequencing run is completed. Setting up this configuration is a simple process that requires just a few clicks, utilizing a secure AWS IAM role and External ID. Customers can apply this configuration to all sequencing runs in their account or selectively to a subset of sequencing runs streaming to a specific S3 bucket. This eliminates the need for customers to develop complex orchestration logic for each individual run.
Upon completion of the workflow, Elembio Cloud enhances the capabilities of Amazon Omics by providing execution records and enabling the visualization of quality control (QC) statistics through an interactive report. From the beginning of the sequencing run to the completion of the analysis, customers maintain full control of their data within their own AWS account. Optionally, customers can allow Element to access statistical metadata for QC run execution, which can then be presented in Elembio Cloud. This setup enables Element to handle the orchestration process while customers retain ownership and control over their data.
Supported pipelines
Element Biosciences offers 3 Bases2Fastq Ready2Run workflows on Amazon Omics. Each workflow is designed to support one of the three sequencing kits, 150Cycles (2×75), 300Cycles (2×150), or 600Cycles, (2×300). Each workflow is priced per run to provide customers with a predictable price for each workflow run.
Amazon Omics also offers various secondary analysis Ready2Run workflows such as Sentieon and NVIDIA Parabricks genomics pipelines to provide customers with a simple solution to align their FASTQ and generate results.
Conclusion
Amazon Omics give customers the ability to run Element’s Bases2Fastq Ready2Run workflow for AVITI and other Ready2Run secondary analysis workflows at scale that are cost-effective, flexible and straightforward sample-to-answer solution, while maintaining end-to-end control of their own data. Amazon Omics provides a consistent, efficient, and scalable platform to perform primary, secondary and tertiary analysis.
To get started with Element Bioscience’s Bases2Fastq Ready2Run workflows, visit the Amazon Omics console.
To learn more about the price for each workflow, visit Amazon Omics Ready2Run workflow pricing.
Authors
Maxim Mass is the Head of Cloud Software at Element Biosciences. He’s developed a passion for building highly usable software backends while working in the San Diego startup space. For the past 10+ years he’s been designing and building cloud-native genomics analyses platforms focusing on ease of use and scalability. He enjoys wilderness camping with his wife and two children, snowboarding, treating guests with home-made pizzas, and travel.
Rosi Bajari is a Staff Product Owner with the Software Product Design team at Element Biosciences. She has 10+ years in NGS & Life Sciences Software industry working to develop innovate and streamlined experiences for genomics data generation and analysis. When not absorbed in solving the next big NGS problem, she spends her free time playing video games, biking, and camping.
Bryan Lajoie is a Principal Scientist with the Software and Bioinformatics team at Element Biosciences. He is currently working with a diverse set of research and clinical customers to build innovative, flexible, and scalable solutions leveraging the accuracy of the AVITI system. He works across a range of Next Generation Sequencing (NGS) applications, with a focus on human disease. He received his PhD from the University of Massachusetts Medical School, under the direction of Job Dekker PhD HHMI, where he worked to develop and pioneer the Hi-C methodology.