AWS for Industries
New capabilities make it easier for healthcare and life science customers to get started, build applications, and scale-up on Amazon Omics
From oncology research to drug discovery to point of care, the unified analysis of various forms of omics data is helping researchers and clinicians generate new insights and offer more personalized care. While the value of multi-omics is apparent, our healthcare and life sciences customers want better tools to get started, build applications, and scale up analysis that will help lower costs and accelerate insights.
We launched Amazon Omics at re:Invent 2022 to help healthcare and life sciences organizations build at-scale to store, query, and analyze genomic, transcriptomic, and other omics data. We’ve already seen customers such as the Children’s Hospital of Philadelphia, Ovation, and G42 Healthcare adopt Amazon Omics to run bioinformatics analysis pipelines at production scale and spend less time managing infrastructure.
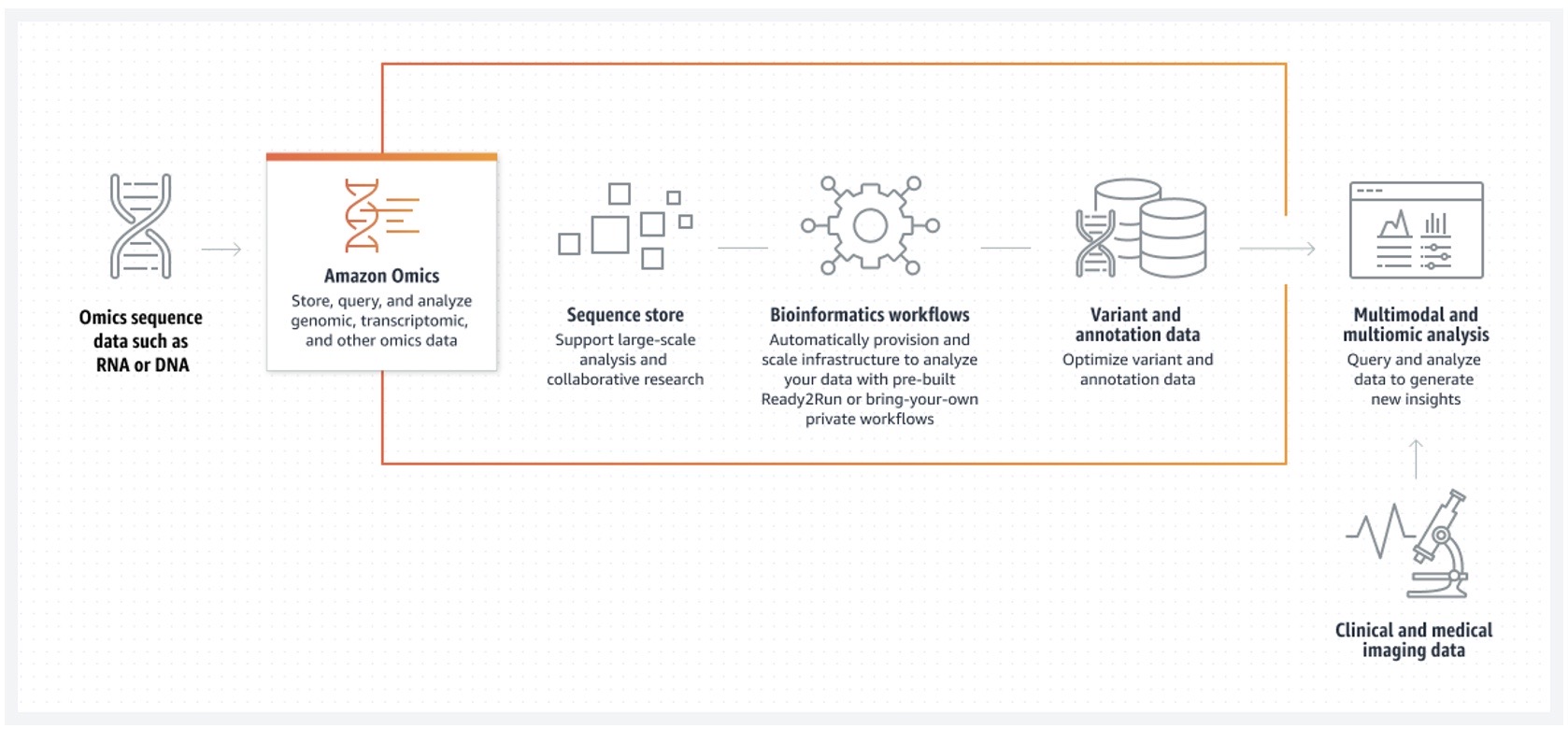 Figure 1. Amazon Omics: How it works across storage, workflows, and analytics
Figure 1. Amazon Omics: How it works across storage, workflows, and analytics
However, there are many use cases where customers want to just run industry standard analysis workflows without any modification. Large-scale analysis of genomic, transcriptomic, and other omics data requires specialized bioinformatics workflows to process data sets. It also requires personnel with a background in biology with knowledge of various programming languages, databases, and tools. Additionally, managing and running these workflows is complex and time-consuming with unpredictable pricing. Doing so locally comes with upfront costs to build and maintain expensive hardware.
To this end, we are excited to announce new capabilities for Amazon Omics designed to make it even easier for healthcare and life sciences organizations to build, run, and scale their workloads. We are thrilled to see these new capabilities are already being used today by customers like Kite Pharma, Columbia University Medical Center, and FYR Diagnostics.
Today at our annual AWS Life Sciences Executive Symposium in Boston, we announced five new capabilities:
- Get started faster: Easily use Ready2Run workflows from Sentieon, NVIDIA Parabricks, Element Biosciences, and open-source pipelines including GATK best practices, nf-core scRNAseq, and AlphaFold and ESMFold for protein prediction
- Accelerated compute power: Ability to use NVIDIA T4 and a10 graphical processing units (GPUs) in Omics workflows to support computationally intensive pipelines
- Direct data ingestion: Directly upload data (FASTQ, CRAM, & BAM) to Omics storage with a new ingestion API
- Easier querying and analysis of variants: Automatically parse variant data containing variant effect predictor (VEP) annotation into a separate data structure for easier querying and analysis
- Event driven applications: Integration with Amazon EventBridge allows customers to now use Amazon Omics published events as part of an event-driven architecture.
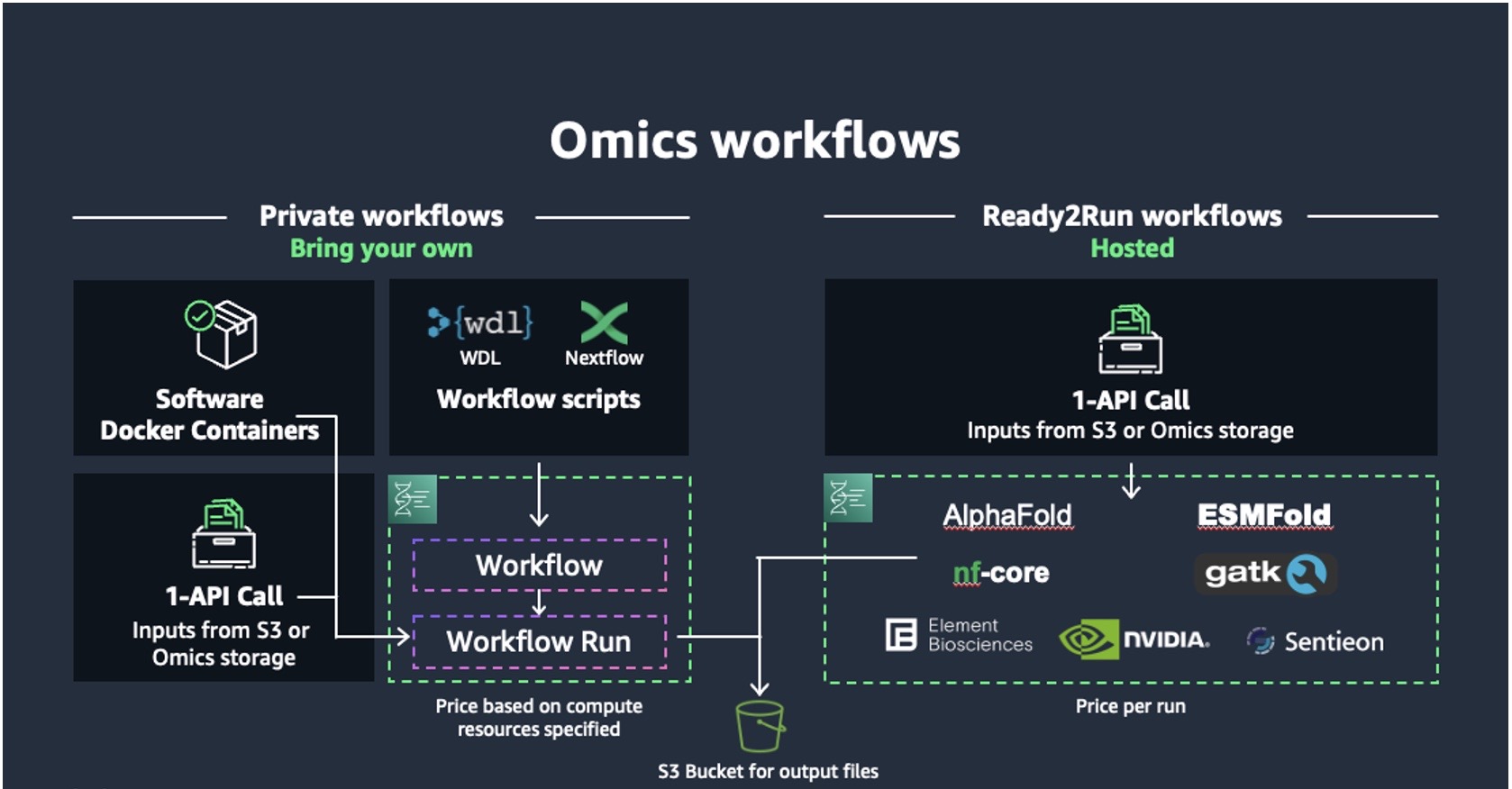 Figure 2. Amazon Omics manages the execution of bioinformatics workflows with private and Ready2Run workflows
Figure 2. Amazon Omics manages the execution of bioinformatics workflows with private and Ready2Run workflows
Get started faster with Ready2Run workflows
Ready2Run workflows are a set of pre-built workflows from third-party software companies and open-source pipelines. With just a few clicks or a single API call, customers can run pre-built pipelines to perform primary analysis such as converting base calls to FASTQ files, secondary analysis such as gene expression or variant calling, and tertiary analysis such as protein structure prediction. Ready2Run workflows are priced-per-run to give customers predictable pricing.
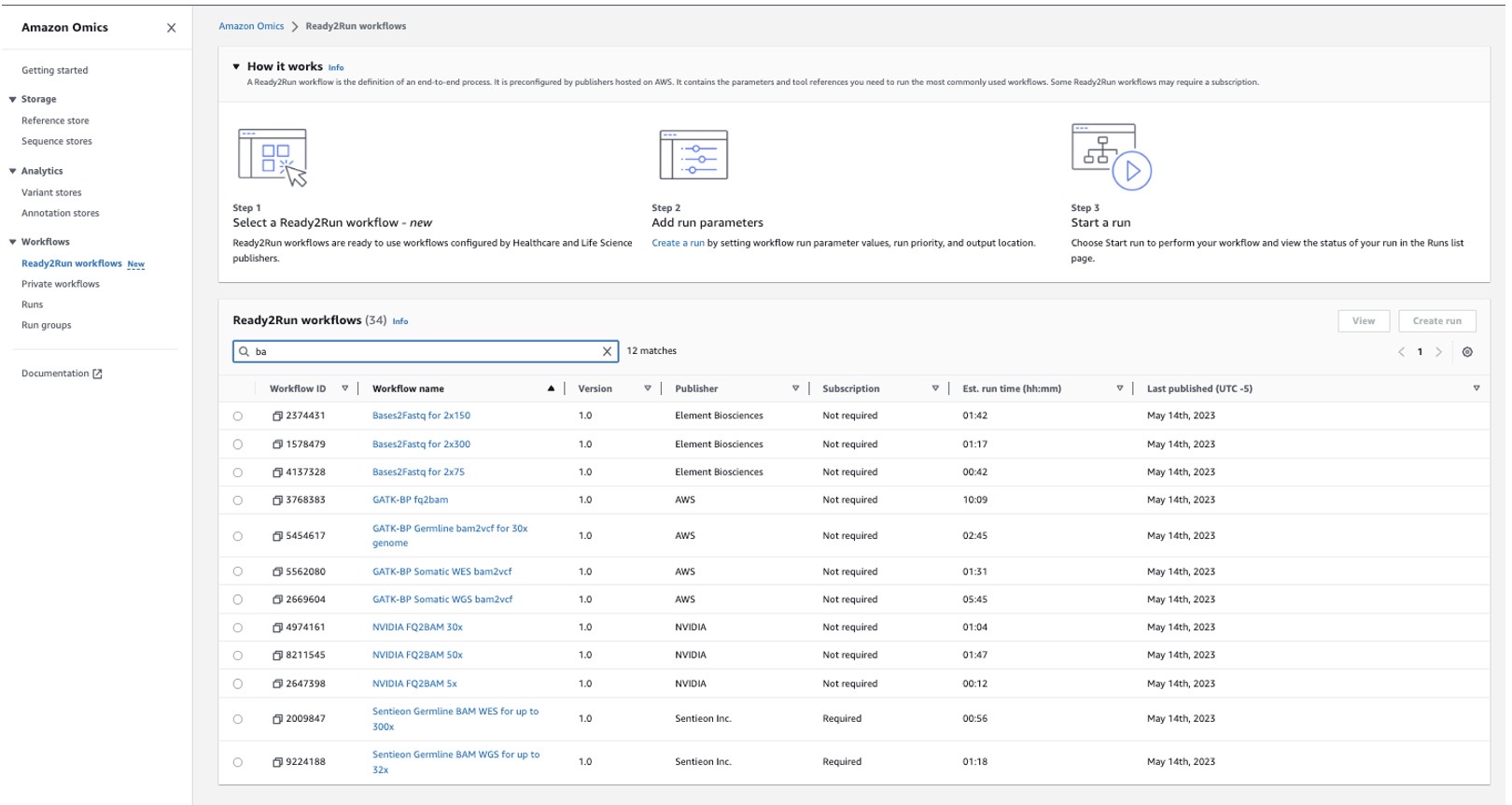 Figure 3. Amazon Omics has 35 pre-built Ready2Run workflows that are easy to search for and select in the console
Figure 3. Amazon Omics has 35 pre-built Ready2Run workflows that are easy to search for and select in the console
With Ready2Run workflows customers have the ability to easily run a set of established pipelines. We are launching with 35 Ready2Run workflows, which are a combination of workflows built by Element Biosciences, NVIDIA, and Sentieon Inc, as well as popular open-source pipelines developed by the life sciences industry.
Customers can now simply bring their data and run these workflows within minutes. They now have access to workflow overviews along with workflow diagrams to give customers detailed information on how the workflow will run in Amazon Omics.
Ready2Run workflows can also be converted into private workflows to provide more flexibility and to support larger total input file sizes for a customer’s specific use case. Private workflows pricing is based on omics instance types and run storage usage.
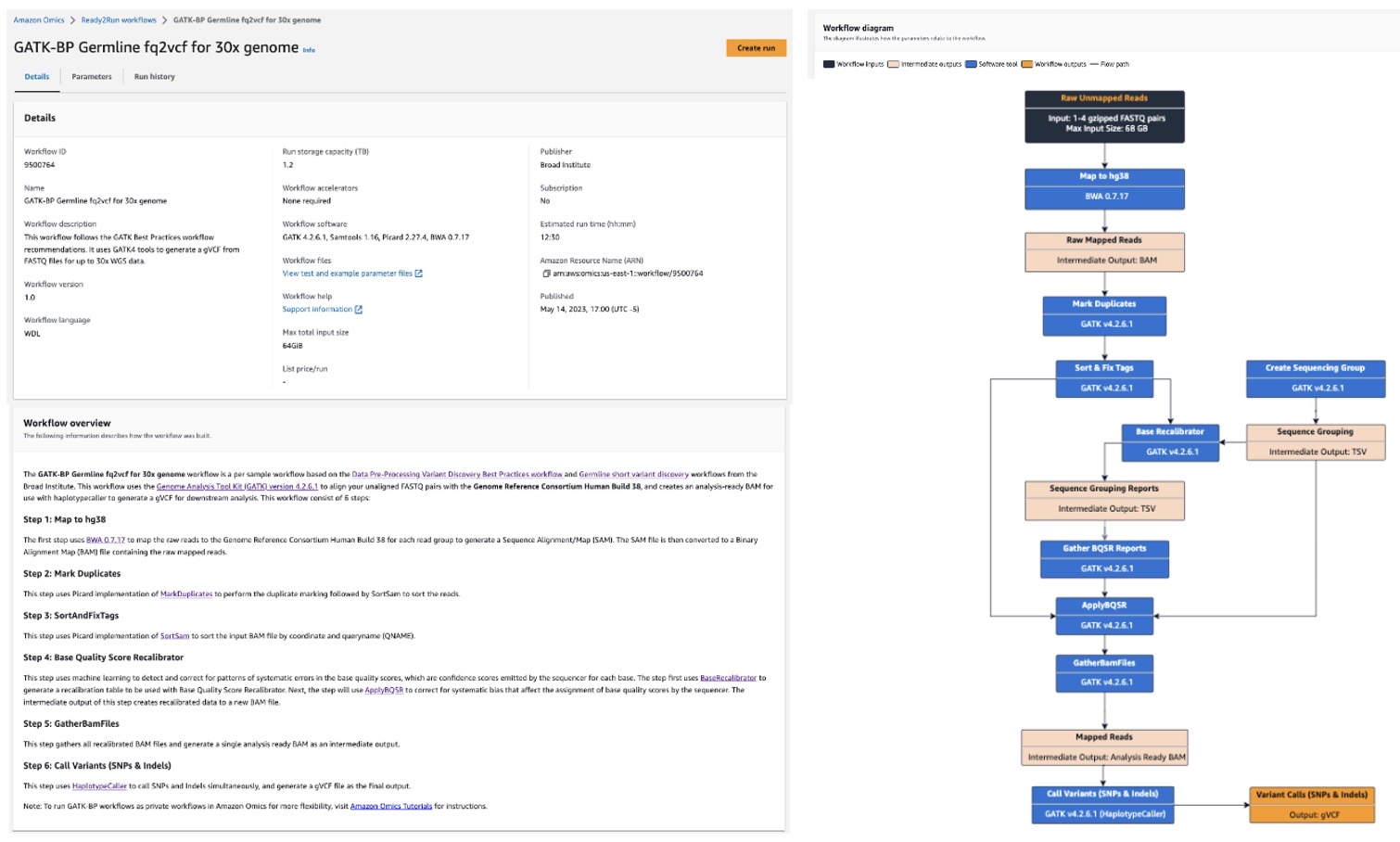 Figure 3. Each Ready2Run workflow has relevant information including description, storage capacity, overview, and diagram
Figure 3. Each Ready2Run workflow has relevant information including description, storage capacity, overview, and diagram
Ready2Run workflows from Sentieon, NVIDIA, and Element Biosciences
Sentieon Ready2Run workflows:
Sentieon workflows provide a robust, scalable, and timely solution for genomic analysis. Amazon Omics offers nine Sentieon Ready2Run workflows including workflows for alignment and germline and somatic variant calling of short-read and long-read datasets. To learn more, read the blog on Sentieon Ready2Run workflows.
NVIDIA Parabricks Ready2Run workflows:
NVIDIA Parabricks is a suite of accelerated genomic analysis applications that contains a variety of optimized and AI-based genomic tools for DNA and RNA, saving costs and time. Amazon Omics offers 13 NVIDIA Parabricks Ready2Run workflows for somatic and germline workflows. To learn more, read the blog on NVIDIA Parabricks workflows.
Element Biosciences Ready2Run workflows:
Element’s Bases2Fastq workflows process the primary data from Element’s AVITI desktop sequencer and generates FASTQ files for use in secondary analysis. Amazon Omics offers Bases2Fastq as a Ready2Run workflow. To learn more, read the blog on Element’s Bases2Fastq workflows.
Open-source Ready2Run workflows
Protein structure prediction workflows:
Protein structure prediction is the process to determine the three-dimensional structure of a protein from its amino acid sequence. The protein structure information enables researchers to better understand the protein’s function, which provides insight into how the protein works, how it interacts with other molecules, and how it can be targeted by drugs.
With DeepMind’s AlphaFold and Meta AI Research’s ESMFold Ready2Run workflows on Amazon Omics, customers can now easily run these workflows to predict the three-dimensional structure of a protein from its amino acid sequence without the need to package the software tools or workflow definition file.
The AlphaFold Ready2Run workflows use the AlphaFold model from DeepMind to predict the 3D structure. The ESMFold Ready2Run workflow uses a large language model from Facebook AI Research (FAIR) to predict the 3D structure. The ESMFold model can predict protein structures up to 60x faster than models based on multiple sequence alignment (MSA) transformers.
GATK Best Practice workflows developed by the Broad Institute:
GATK Best Practices Ready2Run workflows are based on the Data Pre-Processing and Variant Discovery Best Practices from the Broad Institute. These workflows use the Genome Analysis Tool Kit (GATK) version 4.2.6.1.
scRNAseq workflows:
The nf-core scRNAseq Ready2Run workflows are based on the nf-core/scrnaseq pipeline and provide implementation STARsolo, Kallisto, or Salmon Alevin-fry to analyze droplet single cell RNA sequencing data.
Amazon Omics Ready2Run workflow customers
“Kite has a singular focus on cell therapy to treat and potentially cure cancer. We are already seeing the power of Amazon Omics Ready2Run workflows for scRNAseq to analyze single cell RNA sequencing data.” – Jenny Wei, Sr. Director, Head of R&D Informatics & Technology, Kite Pharma, a Gilead Company.
“Columbia University Irving Medical Center (CUIMC) is a clinical, research, and educational enterprise providing global leadership in scientific research, health and medical education, and patient care. I am confident that the ease of use, scalability and cost transparency of AWS’s Ready2Run workflows for GATK Best Practices, NVIDIA Parabricks, and Sentieon’s pipelines significantly lowers barriers to bioinformaticians at any scale. Combined with native integration into AWS’s analytical ecosystem this should significantly accelerate the pace of clinico-genomics.” – Daniel S. T. Hughes PhD, Director of Bioinformatics, Institute for Genomic Medicine & Precision Genomics Laboratory, Columbia University Irving Medical Center
“FYR Diagnostics is developing the next generation of liquid biopsy solutions to meet currently unmet diagnostic needs by harnessing the innovative properties of extracellular vesicles and other revolutionary biomarkers. It was quick and easy to launch the Element Biosciences Bases2Fastq Ready2Run workflow. We are excited to use this solution to streamline our pipelines and significantly shorten the time (and effort) between our data coming off the AVITI and entering secondary analysis for our sequencing service and R&D efforts.” – Claire Seibold, Director of Software and Data Analytics, FYR Diagnostics.
Additional Amazon Omics capabilities
Accelerated compute power with GPU access in private workflows:
Omics private workflows allows customers to bring their own workflow scripts and specify the compute resources that they need for each task in their workflow. Customers can now enable NVIDIA T4 and a10g GPUs for use in Omics private workflows to support accelerated and AI-based genomics analysis with NVIDIA Parabricks and open-source protein folding pipelines.
Direct data ingestion to Omics storage:
Omics storage enables customers to store FASTQ, BAM, and CRAM files at a cost-effective price at scale. Previously Omics had an asynchronous batch upload process for bulk loading of sequence readsets. This new capability adds a simple synchronous upload capability. The multi-part direct upload APIs will now allow customers to easily upload their data directly to the sequence store. This functionality allows customers to integrate existing processing pipelines and/or sequencers to directly write their outputs to a sequence store. Additionally, the transfer manager utility has been updated so that customers can directly upload large files with a single python command.
Easier querying and analysis with automatic variant data parsing:
Omics analytics enables customers to transform Variant Call Files (VCFs) and annotation data for use in multi-omic and multi-modal analysis. With automatic variant data parsing, customers’ VCFs containing annotations generated by the variant effect predictor (VEP) are now separated into a new data structure for easier querying and analysis.
Event-driven applications with Amazon EventBridge integration
Amazon EventBridge is a serverless event bus that makes it easy to connect and route events between AWS services, third-party applications, and customers in their own applications. With Amazon Omics integrated with Amazon EventBridge, customers can now use Amazon Omics published events as part of their event-driven architecture to easily integrate with other AWS services and software applications such as Laboratory Information Management Systems (LIMS) or diagnostic reporting systems.
Conclusion
With the announcement of Ready2Run workflows, GPU support in Omics workflows, direct upload to Omics Storage, automatic variant data parsing, and Amazon EventBridge integration, healthcare and life sciences organizations have new capabilities to help scale their research and accelerate scientific discovery with purpose-built capabilities in Amazon Omics.
Learn more by visiting Amazon Omics.
To get started with Ready2Run workflows visit the Amazon Omics console.
To convert Ready2Run workflows as a private workflow, visit Amazon Omics tutorials.