AWS Big Data Blog
Run Jupyter Notebook and JupyterHub on Amazon EMR
NOTE: Please note that as of EMR 5.14.0, JupyterHub is an officially supported application. We recommend you use the most recent version of EMR if you would like to run JupyterHub on EMR. In addition, EMR Notebooks allow you to create and open Jupyter notebooks with the Amazon EMR console. We will not provide any additional updates to the content in this post.
Tom Zeng is a Solutions Architect for Amazon EMR
Jupyter Notebook (formerly IPython) is one of the most popular user interfaces for running Python, R, Julia, Scala, and other languages to process and visualize data, perform statistical analysis, and train and run machine learning models. Jupyter notebooks are self-contained documents that can include live code, charts, narrative text, and more. The notebooks can be easily converted to HTML, PDF, and other formats for sharing.
Amazon EMR is a popular hosted big data processing service that allows users to easily run Hadoop, Spark, Presto, and other Hadoop ecosystem applications, such as Hive and Pig.
Python, Scala, and R provide support for Spark and Hadoop, and running them in Jupyter on Amazon EMR makes it easy to take advantage of:
- the big-data processing capabilities of Hadoop applications.
- the large selection of Python and R packages for analytics and visualization.
JupyterHub is a multiple-user environment for Jupyter. You can use the following bootstrap action (BA) to install Jupyter and JupyterHub on Amazon EMR:
s3://aws-bigdata-blog/artifacts/aws-blog-emr-jupyter/install-jupyter-emr5.shThese are the supported Jupyter kernels:
- Python
- R
- Scala
- Apache Toree (which provides the Spark, PySpark, SparkR, and SparkSQL kernels)
- Julia
- Ruby
- JavaScript
- CoffeeScript
- Torch
The BA will install Jupyter, JupyterHub, and sample notebooks on the master node.
Commonly used Python and R data science and machine learning packages can be optionally installed on all nodes. Use the Python 2 (or Python 3 if you use --python3 option) notebook to run PySpark code, use the R notebook to run SparkR code, and use Toree Scala notebook to Spark Scala code.
The following arguments can be passed to the BA:
| --r | Install the IRKernel for R. |
| --toree | Install the Apache Toree kernel that supports Scala, PySpark, SQL, SparkR for Apache Spark. |
| --julia | Install the IJulia kernel for Julia. |
| --torch | Install the iTorch kernel for Torch (machine learning and visualization). |
| --ruby | Install the iRuby kernel for Ruby. |
| --ds-packages | Install the Python data science-related packages (scikit-learn pandas statsmodels). |
| --ml-packages | Install the Python machine learning-related packages (theano keras tensorflow). |
| --bigdl | Install Intel’s BigDL deep learning libraries. |
| --python-packages | Install specific Python packages (for example, ggplot and nilearn). |
| --port | Set the port for Jupyter notebook. The default is 8888. |
| --user | Set the default user for JupyterHub, default is jupyter |
| --password | Set the password for the Jupyter notebook. |
| --localhost-only | Restrict Jupyter to listen on localhost only. The default is to listen on all IP addresses. |
| --jupyterhub | Install JupyterHub. |
| --jupyterhub-port | Set the port for JuputerHub. The default is 8000. |
| --notebook-dir | Specify the notebook folder. This could be a local directory or an S3 bucket. |
| --cached-install | Use some cached dependency artifacts on S3 to speed up installation. |
| --ssl | Enable SSL. For production, make sure to use your own certificate and key files. |
| --copy-samples | Copy sample notebooks to the notebook folder. |
| --spark-opts | User-supplied Spark options to override the default values. |
| --python3 | Packages and apps installed for Python 3 instead of Python 2. |
| --s3fs | Use s3fs instead of the default, s3contents for storing notebooks on Amazon S3. This argument can cause slowness if the S3 bucket has lots of files. |
By default (with no --password and --port arguments), Jupyter will run on port 8888 with no password protection; JupyterHub will run on port 8000. The --port and --jupyterhub-port arguments can be used to override the default ports to avoid conflicts with other applications.
The --r option installs the IRKernel for R. It also installs SparkR and sparklyr for R, so make sure Spark is one of the selected EMR applications to be installed. You’ll need the Spark application if you use the --toree argument.
If you used --jupyterhub, use Linux users to sign in to JupyterHub. (Be sure to create passwords for the Linux users first.) jupyter, the default admin user for JupyterHub, can be used to set up other users. The --password option sets the password for Jupyter and for the jupyter user for JupyterHub.
Jupyter on EMR allows users to save their work on Amazon S3 rather than on local storage on the EMR cluster (master node).
To store notebooks on S3, use:
--notebook-dir <s3://your-bucket/folder/>To store notebooks in a directory different from the user’s home directory, use:
--notebook-dir <local directory>The following example CLI command is used to launch a five-node (c3.4xlarge) EMR 5.12. cluster with the bootstrap action. The BA will install all the available kernels. It will also install the ggplot and pybrain Python packages and set:
- the Jupyter port to 8885
- the password to jupyter
- the JupyterHub port to 8005
Replace <your-ec2-key> with your AWS access key and <your-s3-bucket> with the S3 bucket where you store notebooks. You can also change the instance types to suit your needs and budget.
NOTE: If you specify all or most of the options, it will take the bootstrap action longer to run, and on smaller EC2 master instances, it could timeout after 60 minutes, the causing cluster launch to fail. If you’d like to try all the options, we recommend that you use 2xlarge instances (e.g. m3.2xlarge) or higher.
If you are using the EMR console to launch a cluster, you can specify the bootstrap action as follows:
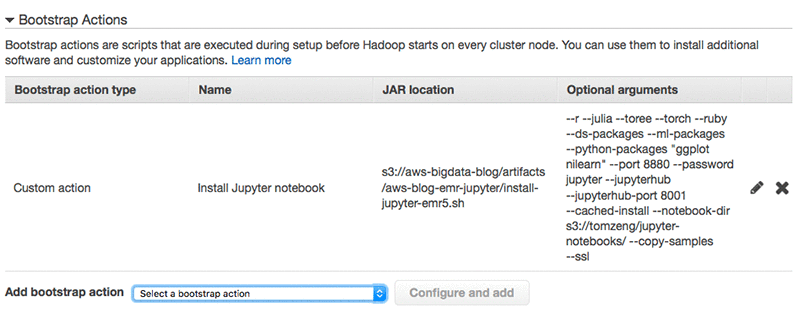
Notice the bootstrap action syntax difference between AWS CLI and AWS EMR Console: the CLI uses comma to separate optional arguments, and the EMR Console uses space. And for the --python-packages argument, the CLI uses single quote for multiple packages. The EMR Console uses double quotes.
When the cluster is available, set up the SSH tunnel and web proxy. The Jupyter notebook should be available at localhost:8885 (as specified in the example CLI command).
After you have signed in, you will see the home page, which displays the notebook files:

If JupyterHub is installed, the Sign in page should be available at port 8005 (as specified in the CLI example):
After you are signed in, you’ll see the JupyterHub and Jupyter home pages are the same. The JupyterHub URL, however, is /user/<username>/tree instead of /tree.
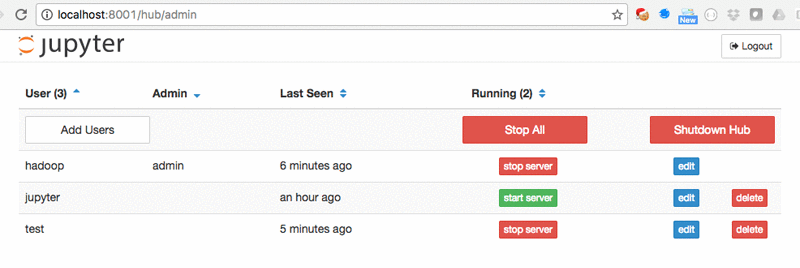
The JupyterHub Admin page is used for managing users:
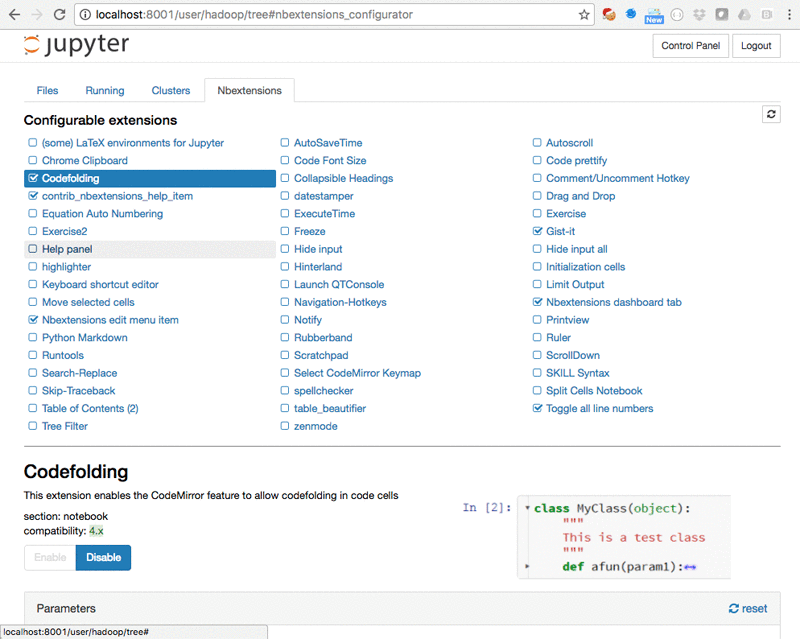
You can install Jupyter extensions from the Nbextensions tab:
If you specified the --copy-samples option in the BA, you should see sample notebooks on the home page. To try the samples, first open and run the CopySampleDataToHDFS.ipynb notebook to copy some sample data files to HDFS. In the CLI example, --python-packages,'ggplot nilearn' is used to install the ggplot and nilearn packages. You can verify those packages were installed by running the Py-ggplot and PyNilearn notebooks.
The CreateUser.ipynb notebook contains examples for setting up JupyterHub users.
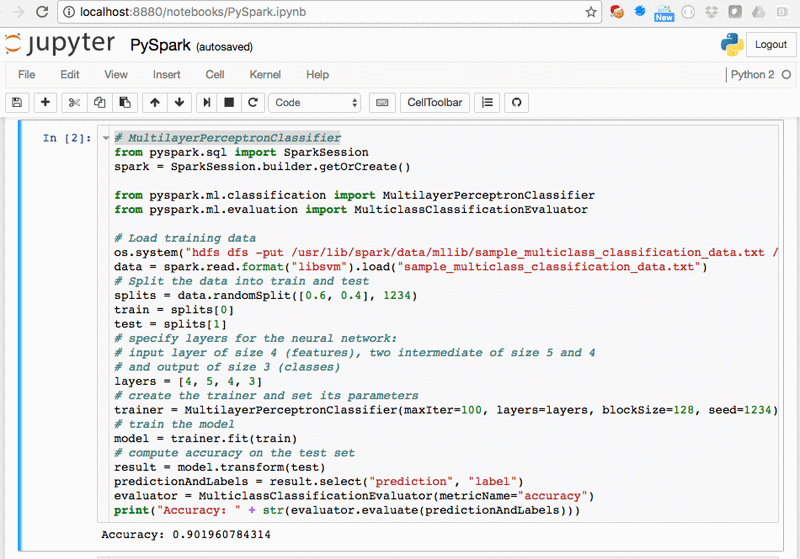
The PySpark.ipynb and ScalaSpark.ipynb notebooks contain the Python and Scala versions of some machine learning examples from the Spark distribution (Logistic Regression, Neural Networks, Random Forest, and Support Vector Machines):
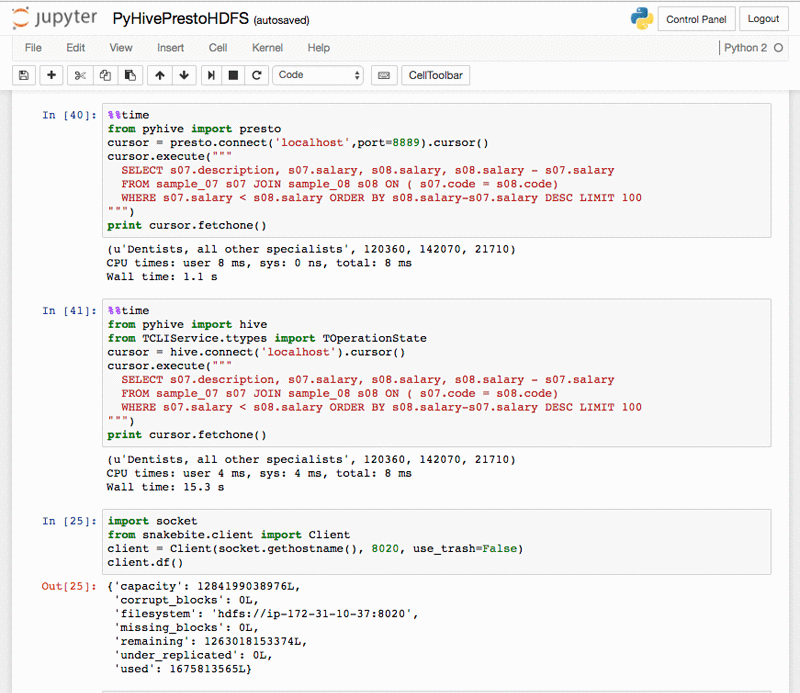
PyHivePrestoHDFS.ipynb shows how to access Hive, Presto, and HDFS in Python. (Be sure to run the CreateHivePrestoS3Tables.ipynb first to create tables.) The %%time and %%timeit cell magics can be used to benchmark Hive and Presto queries (and other executable code):
Here are some other sample notebooks for you to try.
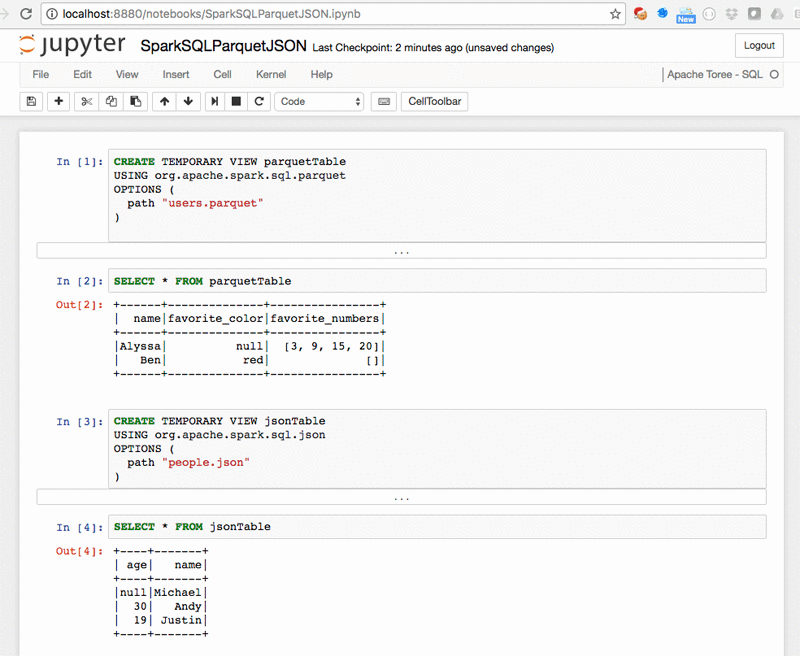
SparkSQL – SparkSQLParquetJSON.ipynb:
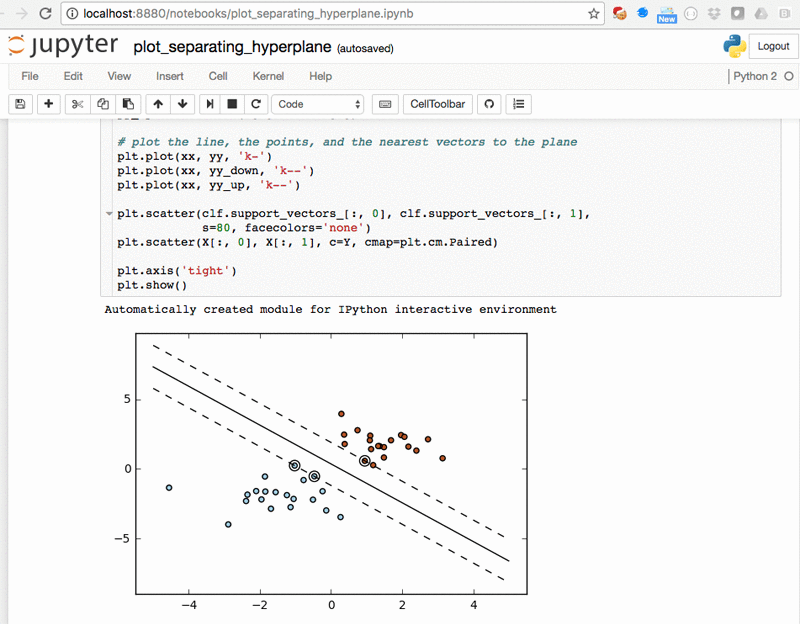
Plot of SVM separating hyperplanes – plot_separating_hyperplane.ipynb:
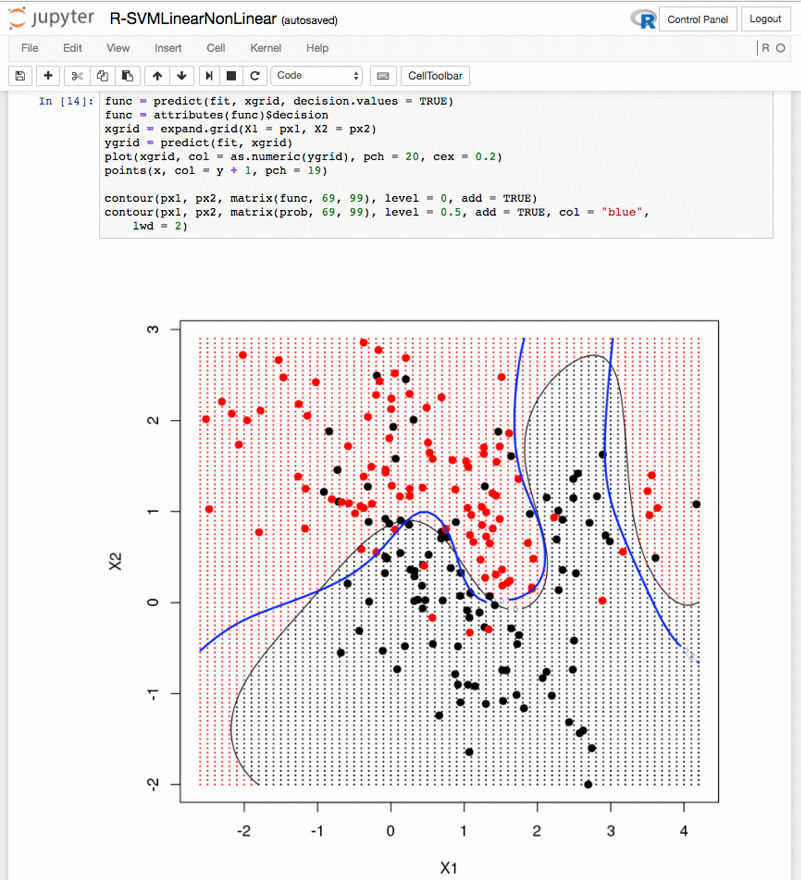
Linear vs non-linear Support Vector Machines – R-SVMLinearNonLinear.ipynb:
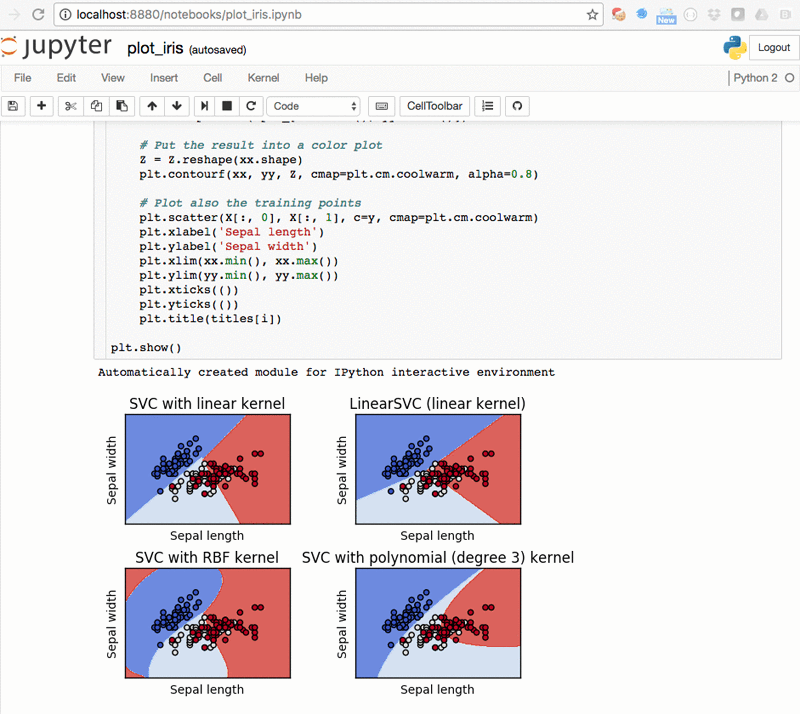
Plots of SVMs in Python – plot_iris.ipynb:
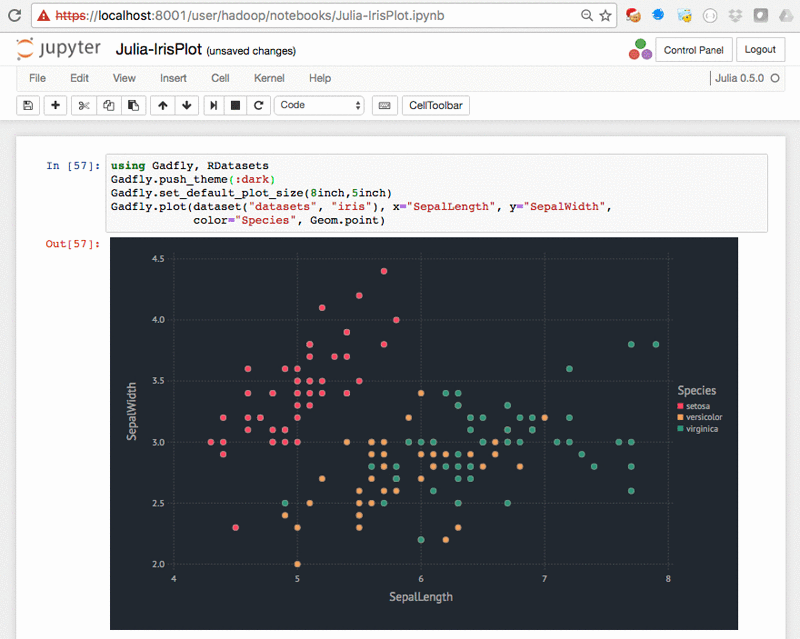
Julia iris plot – Julia-IrisPlot.ipynb:
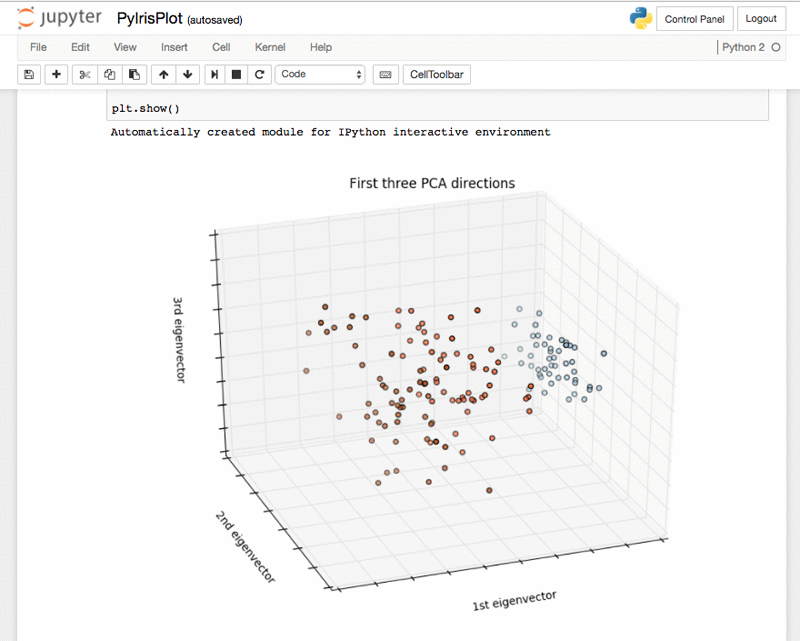
Python iris plot – PyIrisPlot.ipynb:
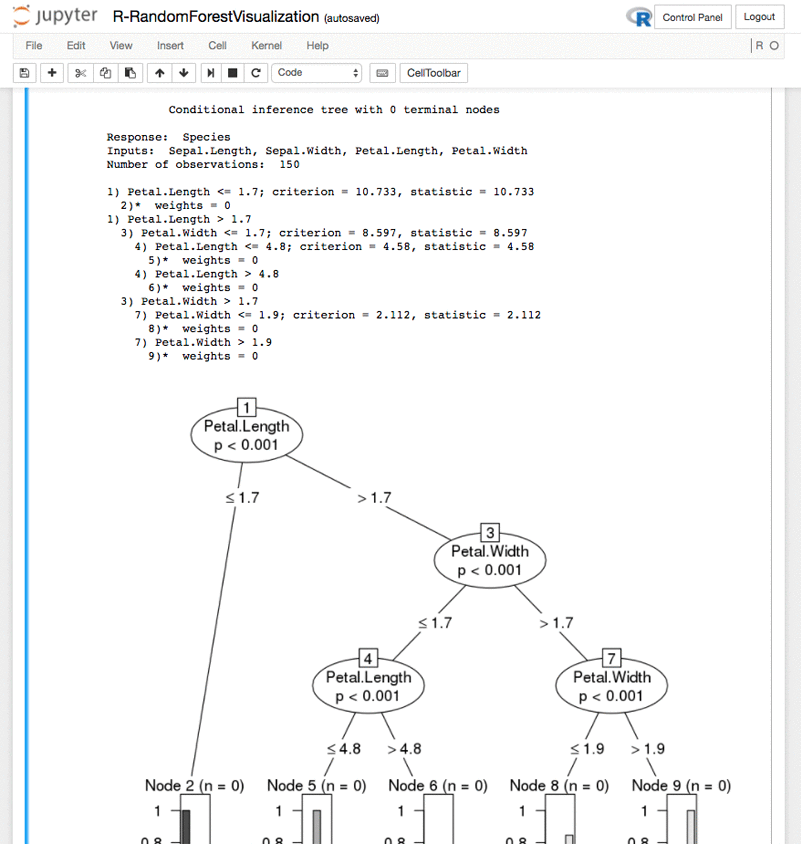
Random Forest tree plot – R-RandomForestVisualization.ipynb:
Granger Causality test in R – GrangerCausality.ipynb:

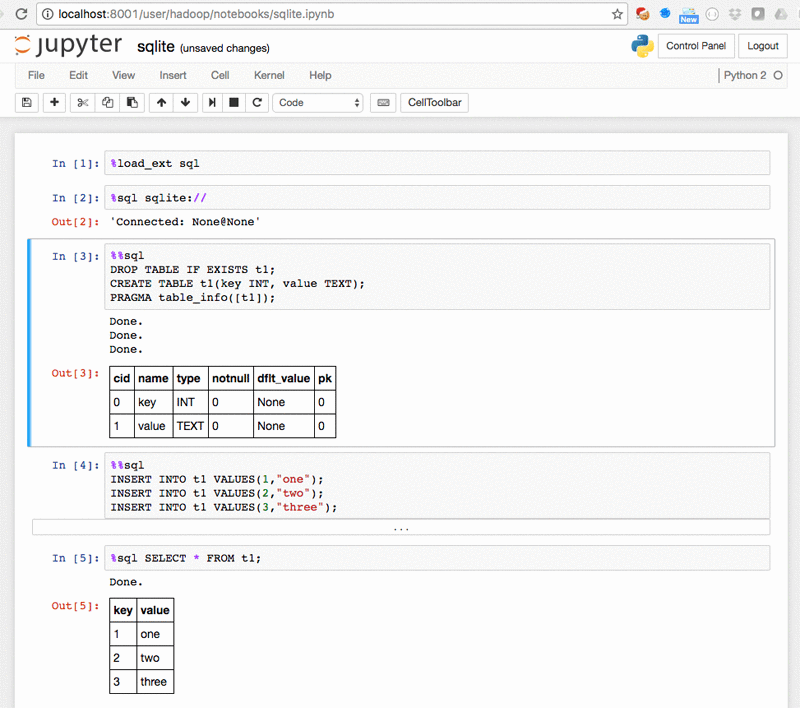
The %%sql cell extension for SQL – SQLite.ipynb:
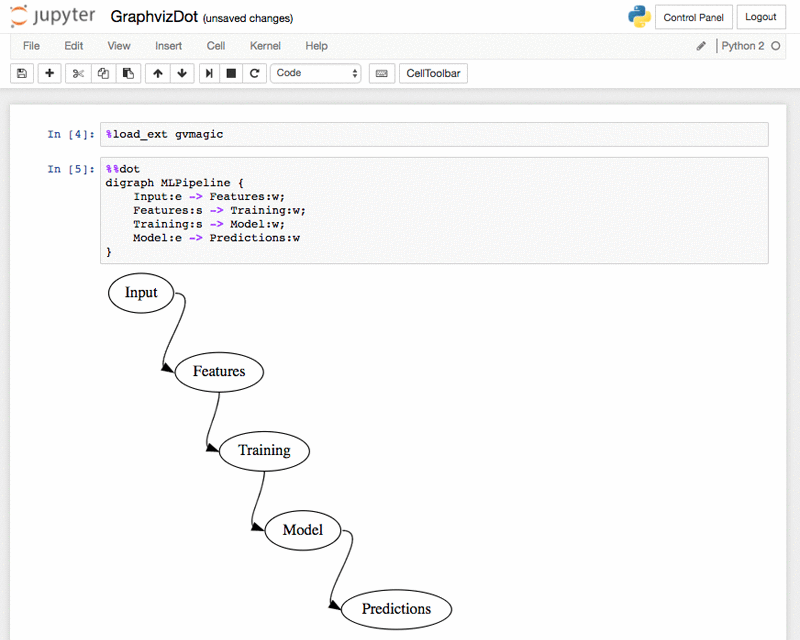
The %%dot extension for Graphviz dot language – GraphvizDot.ipynb:
Conclusion
Data scientists who run Jupyter and JupyterHub on Amazon EMR can use Python, R, Julia, and Scala to process, analyze, and visualize big data stored in Amazon S3. Jupyter notebooks can be saved to S3 automatically, so users can shut down and launch new EMR clusters, as needed. EMR makes it easy to spin up clusters with different sizes and CPU/memory configurations to suit different workloads and budgets. This can greatly reduce the cost of data-science investigations.
If you have questions about using Jupyter and JupyterHub on EMR or would like share your use cases, please leave a comment below.
Related
Running sparklyr – RStudio’s R Interface to Spark on Amazon EMR
