AWS Big Data Blog
Will Spark Power the Data behind Precision Medicine?
Christopher Crosbie is a Healthcare and Life Science Solutions Architect with Amazon Web Services.
This post was co-authored by Ujjwal Ratan, a Solutions Architect with Amazon Web Services.
———————————
“And that’s the promise of precision medicine — delivering the right treatments, at the right time, every time to the right person.“ (President Obama, 2015 State of the Union address)
The promise of precision medicine that President Obama envisions with this statement is a far-reaching goal that will require sweeping changes to the ways physicians treat patients, health data is collected, and global collaborative research is performed. Precision medicine typically describes an approach for treating and preventing disease that takes into account a patient’s individual variation in genes, lifestyle, and environment. Achieving this mission relies on the intersection of several technology innovations and a major restructuring of health data to focus on the genetic makeup of an individual.
The healthcare ecosystem has chosen a variety of tools and techniques for working with big data, but one tool that comes up again and again in many of the architectures we design and review is Spark on Amazon EMR.
Spark is already known for being a major player in big data analysis, but it is additionally uniquely capable in advancing genomics algorithms given the complex nature of genomics research. This post introduces gene analysis using Spark on EMR and ADAM, for those new to precision medicine.
Development of precision medicine
Data-driven tailored treatments have been commonplace for certain treatments like blood transfusions for a long time. Historically, however, most treatment plans are deeply subjective, due to the many disparate pieces of information that physicians must tie together to make a health plan based on the individual’s specific conditions.
To move past the idiosyncratic nature of most medical treatments, we need to amass properly collected and curated biological data to compare and correlate outcomes and biomarkers across varying patient populations.
The data of precision medicine inherently involves the data representation of large volumes of living, mobile, and irrationally complex humans. The recent blog post How The Healthcare of Tomorrow is Being Delivered Today detailed the ways in which AWS underlies some of the most advanced and innovative big data technologies in precision medicine. Technologies like these will enable the research and analysis of the data structures necessary to create true individualized care.
Genomics data sets require exploration
The study of genomics dates back much further than the Obama era. The field benefits from the results of a prodigious amount of research spanning Gregor Mendel’s pea pods in the 1860s to the Human Genome Project of the 1990s.
As with most areas of science, building knowledge often goes hand in hand with legacy analysis features that turn out to be outdated as the discipline evolves. The generation of scientists following Mendel, for example, significantly altered his calculations due to the wide adoption of the p-value in statistics.
The anachronism for many of the most common genomics algorithms today is the failure to properly optimize for cloud technology. Memory requirements are often limited to a single compute node or expect a POSIX file system. These tools may have been sufficient for the computing task involved in analyzing the genome of a single person. However, a shift to cloud computing will be necessary as we move to the full-scale population studies that will be required to develop novel methods in precision medicine.
Many existing genomics algorithms must be refactored to address the scale at which research is done today. The Broad Institute of MIT and Harvard, a leading genomic research center, recognized this and moved many of the algorithms that could be pulled apart and parallelized into a MapReduce paradigm within a Genome Analysis Toolkit (GATK). Despite the MapReduce migration and many Hadoop-based projects such as BioPig, the bioinformatics community did not fully embrace the Hadoop ecosystem due to the sequential nature of the reads and the overhead associated with splitting the MapReduce tasks.
Precision medicine is also going to rely heavily on referencing public data sets. Generally available, open data sets are often downloaded and copied among many researcher centers. These multiple copies of the same data create an inefficiency for researchers that is addressed through the AWS Public Data Sets program. This program allows researchers to leverage popular genomics data sets like TCGA and ICGC without having to pay for raw storage.
Migrating genetics to Spark on Amazon EMR
The transitioning of genomics algorithms that are popular today to Spark is one path that scientists are taking to capture the distributed processing capabilities of the cloud. However, precision medicine will require an abundance of exploration and new approaches. Many of these are already being built on top of Spark on EMR.
This is because Spark on EMR provides much of the functionality that aligns well with the goals of the precision medicine. For instance, using Amazon S3 as an extension of HDFS storage makes it easy to share the results of your analysis with collaborators from all over the world by simply allowing access to an S3 URL. It also provides the ability for researchers to adjust their cluster to the algorithm they are trying to build instead of adjusting their algorithm to the cluster to which they have access. Competition for compute resources with other cluster users is another drawback that can be mitigated with a move towards EMR.
The introduction of Spark has now overcome many of the previous limitations associated with parallelizing the data based on index files that comprise the standard Variant Call Format (VCF) and Binary Alignment Map (BAM) formats used by genomics researchers. The AMPLab at UC Berkeley, through projects like ADAM, demonstrated the value of a Spark infrastructure for genomics data processing.
Moreover, genomics scientists began identifying the need to get away from the undifferentiated heavy lifting of developing custom distributed system code. These developments motivated the Broad to develop the next version of the GATK with an option to run in the cloud on Spark. The upcoming GATK is a major step forward for the scientific community since it will soon be able to incorporate many of the features of EMR, such as on-demand cluster of various types and Amazon S3–backed storage.
Although Spark on EMR provides many infrastructure advantages, Spark still speaks the languages that are popular with the research community. Languages like SparkR on EMR make for an easy transition into the cloud. That way when there are issues that arise such as needing to unpack Gzip-compressed files to make them split-able, familiar R code can be used to make the transformation.
What’s under the hood of ADAM?
ADAM is an in-memory MapReduce system. ADAM is the result of contributions from universities, biotech, and pharma companies. It is entirely open source under the Apache 2 license. ADAM uses Apache Avro and Parquet file formats for data storage to provide a schema-based description of genomic data. This eliminates the dependencies on format-based libraries, which in the past has created incompatibilities.
Apache Parquet is a columnar storage format that is well suited for genomic data. The primary reason for this is because genomic data consists of a large number of similar data items that is align well with columnar storage. By using Apache Parquet as its storage format, ADAM makes querying and storing genomic data highly efficient. It limits the I/O to data actually needed by loading only the columns that need to be accessed. Because of its columnar nature, storing data in Parquet saves space as a result of better compression ratios.
The columnar Parquet file format enables up to 25% improvement in storage volume compared to compressed genomics file formats like BAM. Moreover, the in-memory caching using Spark and the ability to parallelize data processing over multiple nodes have been known to provide up to 50 times better performance on average on a 100-node cluster.
In addition to using the Parquet format for columnar storage, ADAM makes use of a new schema for genomics data referred to as bdg-formats, a project that provides schemas for describing common genomic data types such as variants, assemblies, and genotypes. It uses Apache Avro as its base framework and as a result, works well with common programming languages and platforms. By using the Apache Avro-based schema as its core data structure, workflows built using ADAM are flexible and easy to maintain using familiar technologies.
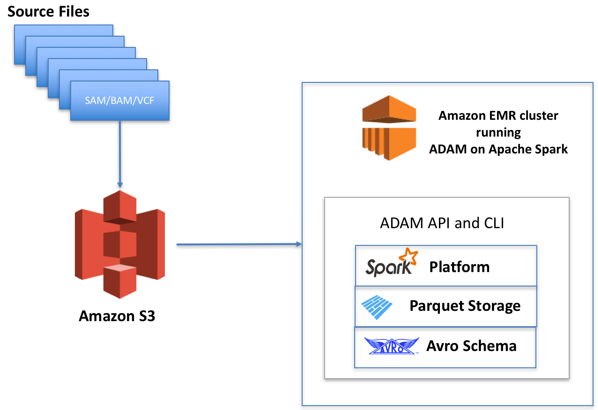
ADAM on Amazon EMR
The figure above represents a typical genomic workload designed on AWS using ADAM. File formats like SAM/BAM and VCF are uploaded as objects on Amazon S3.
Amazon EMR provides a way to run Spark compute jobs with ADAM and other applications on an as-needed basis while keeping the genomics data itself stored in separate, cheap object storage.
Your first genomics analysis with ADAM on Amazon EMR
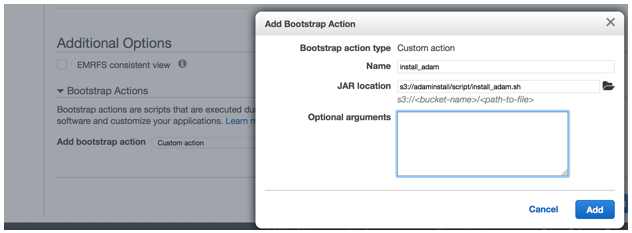
To install ADAM on EMR, launch an EMR cluster from the AWS Management Console. Make sure you select the option to install Spark, as shown in the screen shot below.

While launching the EMR cluster, you should configure the EMR master security groups to allow you access to port 22 so you can SSH to the master node. The master node should be able to communicate to the Internet to download the necessary packages and build ADAM.
ADAM installation steps
During the creation of a cluster, a shell script known as a bootstrap action can automate the installation process of ADAM. In case you want to install ADAM manually, the following instructions walk through what the script does.
After the EMR cluster with Apache Spark is up and running, you can log in into the master node using SSH and install ADAM. ADAM requires Maven to build the packages; after you install Maven, you can clone ADAM from the ADAM GitHub repository.
- SSH into the master node of the EMR cluster.
- Install Maven by downloading the apache libraries from its website as shown below:
/* create a new directory to install maven */ mkdir maven cd maven /* download the maven zip from the apache mirror website */ echo "copying the maven zip file from the apache mirror" wget http://apachemirror.ovidiudan.com/maven/maven-3/3.3.9/binaries/apache-maven-3.3.9-bin.tar.gz echo "unzipping the file" tar -zxvf apache-maven-3.3.9-bin.tar.gz /* export the “MAVEN_HOME” path */ echo "exporting MAVEN_HOME" export PATH=$HOME/maven/apache-maven-3.3.9/bin:$PATH
- Before cloning ADAM from GitHub, install GIT on the the master node of the EMR cluster. This can be done by running the following command:
/* install git in the home directory */ cd $HOME sudo yum install git
- Clone the ADAM repository from GitHub and begin your build.
/* clone the ADAM repository from GitHub and build the package */ git clone https://github.com/bigdatagenomics/adam.git cd adam
- Finally, begin your build
export MAVEN_OPTS="-Xmx512m -XX:MaxPermSize=256m" mvn clean package -DskipTests
- On completion, you see a message confirming that ADAM was built successfully.
Analysis of genomic data using ADAM
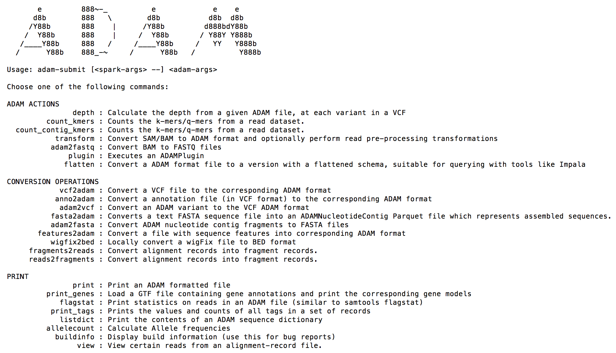
After installation, ADAM provides a set of functions to transform and analyze genomic data sets. The functions range from actions to count K-mers to conversion operations to convert file standard genomics formats like SAM, BAM or VCF to ADAM Parquet. A list of ADAM functions can be viewed by invoking adam-submit from the command line.
For the purposes of this analysis, you use a VCF file from an AWS public data set, specifically the 1000 genome project.
This is a project that aims to build the most detailed map of human genetic variation available. When complete, the 1000 genome project will have genomic data from over 2,661 people. Amazon S3 hosts the initial pilot data for this project in a public S3 bucket. The latest data set available to everyone hosts data for approximately 1700 people and is more than 200 TB in size.
From the master node of the EMR cluster, you can connect to the S3 bucket and download the file to the master node. The first step is to copy the file into HDFS so it can be accessed using ADAM. This can be done by running the following commands:
//copying a vcf file from S3 to master node $ aws s3 cp s3://1000genomes/phase1/analysis_results/integrated_call_sets/ALL.chr1.integrated_phase1_v3.20101123.snps_indels_svs.genotypes.vcf.gz /home/hadoop/ // Unzip the file $ gunzip ALL.chr1.integrated_phase1_v3.20101123.snps_indels_svs.genotypes.vcf.gz //copying file to hdfs $ hadoop fs -put /home/hadoop/ ALL.chr1.integrated_phase1_v3.20101123.snps_indels_svs.genotypes.vcf /user/hadoop/
Once done, you can use the function vcf2adam to convert the vcf file into multiple zipped adam parquet files.
// Converting the file from VCF to adam parquet $ adam-submit vcf2adam "/user/hadoop/ALL.chr1.integrated_phase1_v3.20101123.snps_indels_svs.genotypes.vcf" "/user/hadoop/adamfiles”
The command generates more than 690 ADAM Parquet files. The following screenshot shows a subset of these files.
You can now process the ADAM Parquet files as regular Parquet files in Apache Spark using Scala. ADAM provides its users its own shell, which is invoked from the command line using adam-shell. In this example, read the ADAM Parquet file as a data frame and print its schema. Then, go on to register the data frame as a temp table and use it to query the genomic data sets.
val gnomeDF = sqlContext.read.parquet("/user/hadoop/adamfiles/ ")
gnomeDF.printSchema()
gnomeDF.registerTempTable("gnome")
val gnome_data = sqlContext.sql("select count(*) from gnome")
gnome_data.show()
Here’s how to use ADAM interactive shell to perform a reads count and K-mer count. A k-mer count returns all of the strings of length k from this DNA sequence. This is an important first step to understanding all of the possible DNA sequences (of length 20) that are contained in this file.
import org.bdgenomics.adam.rdd.ADAMContext._
val reads = sc.loadAlignments("part-r-00000.gz.parquet").cache()
reads.count()
val gnomeDF = sqlContext.read.parquet("part-r-00000.gz.parquet")
gnomeDF.printSchema()
println(reads.first)
/* The following command cuts reads into _k_-mers, and then counts the number of occurrences of each _k_-mer */ val Kmers = reads.adamCountKmers(20).cache() kmers.count()
Conclusion
Spark on Amazon EMR provides unique advantages over traditional genomic processing and may become a necessary tool as genomics moves into the scale of population-based studies required for precision medicine. Innovative companies like Human Longevity Inc. have discussed at re:Invent how they use AWS tools such as Spark on EMR with Amazon Redshift to build a platform that is pushing precision medicine forward.
Of course, simply distributing compute resources will not solve all of the complexities associated with understanding the human condition. There is an old adage that reminds us that nine women cannot be used to make a baby in one month. Often in biology problems, there is a need to wait for one step to be completed before the next can be undergone. This procedure does not necessarily lend itself well to Spark, which benefits from distributing many small tasks at once. There are still many areas such as sequence assembly that may not have an easy transition to Spark. However, Spark on Amazon EMR will be a very interesting project to watch on our move toward precision medicine.
If you have questions or suggestions, please leave a comment below.
Special thanks to Angel Pizarro for his help on this post!