AWS News Blog
New AWS Public Data Sets – TCGA and ICGC
|
|
My colleagues Angel Pizarro and Ariel Gold wrote the incredibly interesting guest post below.
— Jeff;
Today we are pleased to announce that qualified researchers can now access two of the world’s largest collections of cancer genome data at no cost on AWS as part of the AWS Public Data Sets program. Providing access to these petabyte-scale genomic data as shared resources on AWS lowers the barrier to entry, thus expanding the research community and accelerating the pace of research and discovery in the development of new treatments for cancer patients.
The Cancer Genome Atlas (TCGA) corpus of raw and processed genomic, transcriptomic, and epigenomic data from thousands of cancer patients is now freely available on Amazon S3 to users of the Cancer Genomics Cloud, a cloud pilot program funded by the National Cancer Institute and powered by the Seven Bridges Genomics platform.
The International Cancer Genome Consortium (ICGC) PanCancer dataset generated by the Pancancer Analysis of Whole Genomes (PCAWG) study is also now available on AWS, giving cancer researchers access to over 2,400 consistently analyzed genomes corresponding to over 1,100 unique ICGC donors. These data will also be freely available on Amazon S3 for credentialed researchers subject to ICGC data sharing policies.
These two data sets represent the first controlled-access genomic data that have been redistributed to the wider research audience on the cloud. Previously, researchers needed to download and store their own copies of the data before they could begin their experiments. Now, with this data hosted on AWS for the community, researchers can begin their work right away. Researchers will also have access to a broader toolset hosted and shared by the community within AWS. This translates into a much lower barrier to entry and more time for science.
Making these data and tools available in the cloud will also enable a greater level of collaboration across research groups, since they will have a common place to access and share data. Finally, researchers will also be able to securely bring their own data and tools into AWS, and combine these with the existing public data for more robust analysis. No-cost data access, a broader set of available tools, and increased collaborative capabilities will enable researchers to focus on their science and not infrastructure, allowing them to get more done in shorter periods, and ultimately accelerating the pace of research and discovery in the study of cancer.
Accessing TCGA and ICGC on AWS
The difference between TCGA and ICGC, and previously released AWS Public Data Sets such as the National Institutes of Health (NIH) 1000 Genomes Project, Genome in a Bottle (GIAB), and the 3000 Rice Genome, is the need to limit access to researchers that have gone through a review process for their intended use of the data. Because of this requirement, access to TCGA and ICGC on AWS will be administered by our third-party partners, Seven Bridges Genomics and the Ontario Institute for Cancer Research, respectively. These partners have the rights to redistribute the data on behalf of the original data providers. The partners will also curate and update the data over time, as well as develop a community of users who can share cloud-based tools and best practices in order to accelerate use of the data and advance our understanding of cancer.
You can learn more about the data sets, and specifics on how to access them, on our TCGA on AWS page and ICGC on AWS page.
Tools and Resources for Working with the Data
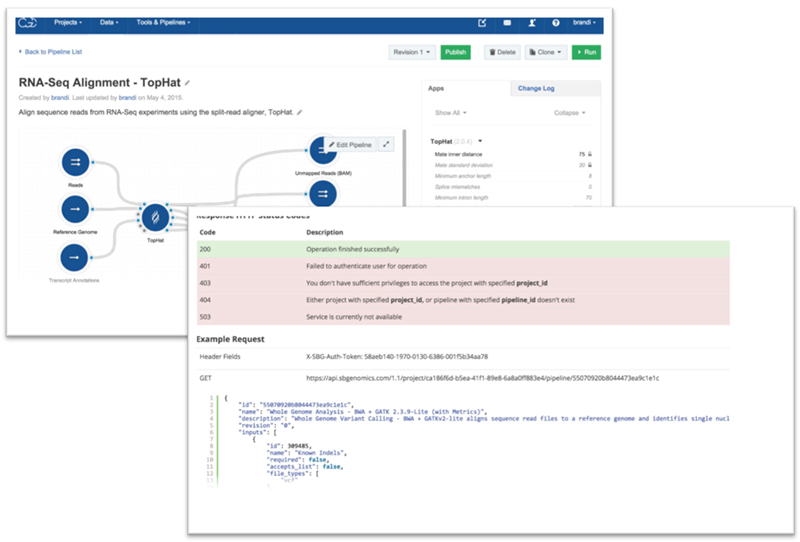
The TCGA data will be available to users of the Cancer Genomics Cloud (CGC). Researchers can apply for early access here. Once accepted, users will be able access the data via the CGC Web portal or use the CGC’s API for programmatic access to the data. The CGC will have a set of data analysis pipelines already integrated into the platform so that users can start working right away with the most common toolsets.
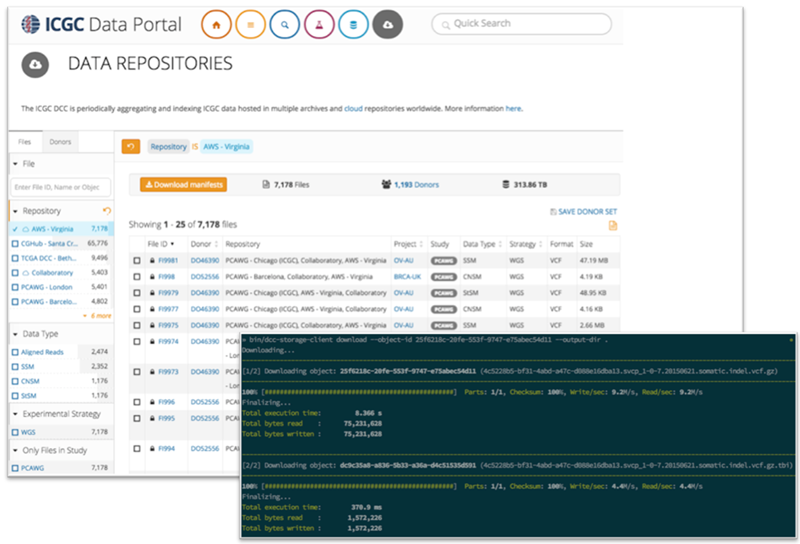
 The ICGC data will be generally accessible via the use of a downloadable command line tool. Users can search for files using the ICGC Data Portal and access individual or related sets of alignment and variant files through the ICGC Storage Client. The alignments and a selection of Sanger somatic variant calls are currently available in Amazon S3. Further variant calls will be released following additional quality checking, validation, and analysis. For more information see the ICGC on the Cloud page and ICGC Storage Client documentation.
The ICGC data will be generally accessible via the use of a downloadable command line tool. Users can search for files using the ICGC Data Portal and access individual or related sets of alignment and variant files through the ICGC Storage Client. The alignments and a selection of Sanger somatic variant calls are currently available in Amazon S3. Further variant calls will be released following additional quality checking, validation, and analysis. For more information see the ICGC on the Cloud page and ICGC Storage Client documentation.
As always, when working with sensitive genomics data on AWS, you should take care to secure your storage and computational resources. The Architecting for Genomic Data Security and Compliance in AWS whitepaper is a good starting point if you are unfamiliar with the service features and tools necessary to work with data in a secure manner. Genomics platforms such as the CGC take care to meet these types of requirements as part of their value proposition. For example, DNAnexus has provided user documentation on how to leverage the ICGC Storage Client within their platform here.
Recognizing that it is no easy task to work with data at this scale, the PCAWG group are also releasing the PanCancer Launcher. This is an open source system to create EC2 instances, enqueue the analysis work items, trigger Docker-based analysis pipelines, and clean up the launched resources as computational tasks complete.
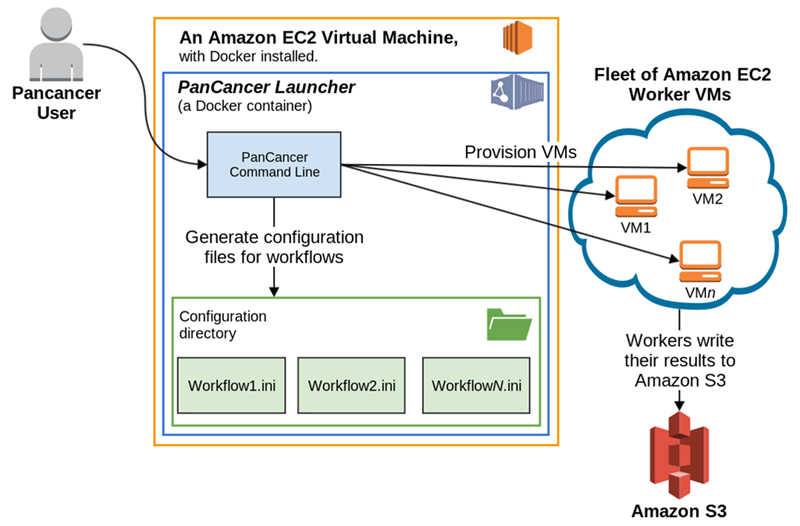
Currently, the PanCancer Launcher includes support for the BWA-mem-based alignment pipeline and its associated quality control steps. Future releases will expand support for the variant calling pipelines created by the project that encompassed current best practice variant calling pipelines from 4 academic organizations: the German Cancer Research Center (DKFZ), the European Molecular Biology Laboratory (EMBL) in Heidelberg, the Wellcome Trust Sanger Institute, and the Broad Institute. You can read more about how to leverage the PanCancer Launcher in the Launcher HOWTO Guide.
Genomics in the Era of Cloud Computing
It has been interesting to witness the parallel evolution of genomics and cloud computing over the last decade. Both have been driven by new technologies that leverage economies of scale. Both have fundamentally changed the types of questions that can be asked simply because we can now collect and analyze the data in the same place.
The genomics research community, which have witnessed their storage and compute requirements double overnight when new chemistry kits are released, realized long ago that scalable cloud computing models are a better fit than large capital purchases that have to be planned for and amortized over 3-5 years. Today, it is common practice to work with data sets that reach in the hundreds of terabytes, and a few important ones that reach into the petabytes like the TCGA and ICGC. For genomics, cloud has become the new normal for how science gets done.
You can learn more about how genomics thought leaders are innovating in the genomics field through the use of cloud in this new video:
Be sure to also visit the Scientific Computing on AWS and Genomics on AWS pages for more user stories and tools.
Thank You
We’d like to thank our collaborators at the Ontario Institute for Cancer Research and Seven Bridges Genomics who helped us launch these public data sets and will be curating the data, administering access, and cultivating the ecosystem of tools around them. We look forward to working with many more organizations and researchers who will share their expertise and tools in order to accelerate the development of new treatments for cancer patients. Tell us how you’re using the data via the TCGA on AWS and ICGC on AWS pages and sign up for project updates.
— Angel Pizarro (AWS Scientific Computing) and Ariel Gold (AWS Public Data Sets)

