AWS HPC Blog
Evaluating next‑generation cloud compute for large‑scale genomic processing
This post was contributed by Manu Pillai (AWS), Marissa E Powers PhD (AWS), Sean O’Dell (AstraZeneca), Gabriel Hernandez (AstraZeneca), Heejoon Jo (Illumina), Shyamal Mehtalia (Illumina), Joe Warren (AWS), and Natalia Jimenez PhD (AWS).
Genomic research demands massive computational power to process and analyze DNA sequences that unlock insights into human health and disease. For pharmaceutical companies, processing millions of genomes efficiently and sustainably is a crucial factor to accelerate discovery and development of future therapies that have the potential to help people live longer, healthier lives.
As cloud infrastructure evolves, many organizations face the challenge of migrating to newer, more powerful instances while maintaining research integrity and managing costs. When Amazon Elastic Compute Cloud (Amazon EC2) F1 instances approach end-of-service in 2025, organisations relying on Field-Programmable Gate Array (FPGA)-based instances to process genetic data may need a clear path forward that doesn’t compromise critical workloads.
In this post, we describe a structured evaluation of Amazon EC2 F2 instances for accelerated genomics workloads, focusing on performance, cost, and output equivalency relative to earlier FPGA-based instance generations.
The challenge: future-proofing genomic infrastructure
AstraZeneca’s Centre for Genomics Research (CGR) integrates large-scale genomic and clinical data leveraging multi-omics approaches—genomics, transcriptomics, proteomics, and metabolomics—at population scale, making it one of the largest and most diverse datasets globally.
The CGR use Illumina DRAGEN®[1] (Dynamic Read Analysis for GENomics) workloads for processing Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) data. With F1 instances approaching end-of-service, we evaluated F2 instances as the next-generation solution for continuing workloads without disruption.
The solution: comprehensive F1 to F2 migration testing
In collaboration with AstraZeneca and Illumina, AWS led extensive testing comparing F1 and F2 instance performance across multiple genomic workloads and AWS Regions.
Testing methodology
The team designed a comprehensive evaluation using:
- Sample Analysis: 11 genomic samples (5 WES and 6 WGS) representing typical research workloads
- Architecture: AWS Batch with separate compute queues for F1 and F2 instances
- Illumina DRAGEN Versions: v3.7.8 for WGS processing, v4.3.6 for WES processing
- Multi-Region Validation: Testing across three AWS regions to ensure global scalability
- Performance Metrics: Runtime comparison and cost analysis for identical workloads
- Validation: A formal method for validating genomic equivalency
The architecture leveraged the Nextflow workflow system with AWS Batch (a fully managed batch computing service) to orchestrate DRAGEN pipeline execution, enabling consistent testing conditions across both instance types.
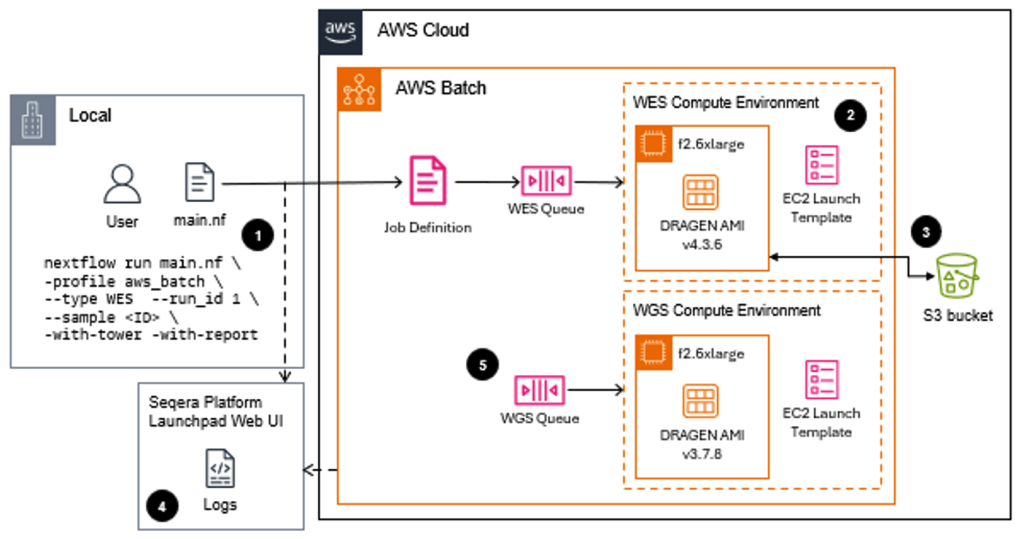
Figure 1 – This diagram shows the cloud testing environment used to compare F1 and F2 instance performance for genomic processing. Users submit DRAGEN workflows through Nextflow, which orchestrates separate AWS Batch compute environments for Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) samples on F2.6xlarge instances. The standardized architecture enabled precise performance comparisons by maintaining identical workflow orchestration while only changing the underlying compute infrastructure, ensuring the 60% performance improvements and 70% cost reductions could be accurately attributed to F2 instances.
Results: dramatic performance and cost improvements
The testing revealed significant improvements when migrating from F1 to F2 instances:
Key Benefits
- 60% faster processing: Significant reduction in genomic analysis runtime
- 70% cost reduction: Substantial savings on compute costs
- Sustainability improvement: Reduced energy use and associated carbon emissions by completing workloads more efficiently
- Enhanced scalability: Improved performance across multiple AWS regions
- Maintained accuracy: No compromise in research data integrity
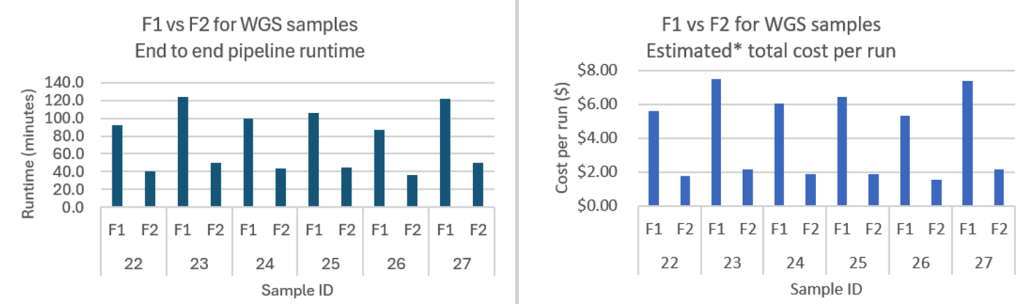
Figure 2 – This chart compares F1.4xlarge and F2.6xlarge instance performance for six Whole Genome Sequencing samples, showing both runtime (in minutes) and cost per run (in dollars). F2 instances consistently deliver up to 60% faster processing times and 70% lower costs across all samples, demonstrating the substantial performance improvements and cost savings organizations can achieve when upgrading from F1 to F2 instances for genomic workloads. On Demand pricing for f1.4xlarge in eu-west-1 and f2.6xlarge in eu-west-2 as of 9/16/2025.
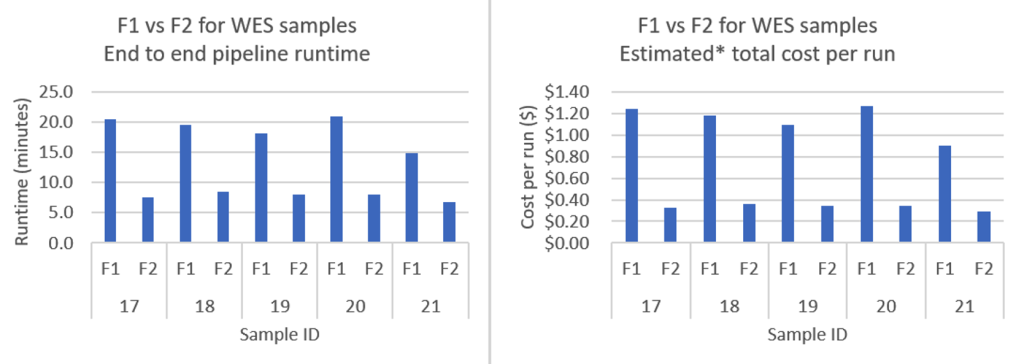
Figure 3 – This chart compares F1.4xlarge and F2.6xlarge instance performance for five Whole Exome Sequencing samples, showing both runtime (in minutes) and cost per run (in dollars). F2 instances consistently deliver up to 63% faster processing times and 74% lower costs across all samples, demonstrating the substantial performance improvements and cost savings organizations can achieve when upgrading from F1 to F2 instances for exome sequencing workloads. On Demand pricing for f1.4xlarge in eu-west-1 and f2.6xlarge in eu-west-2 as of 9/16/2025.
Validating equivalency
To assess Illumina DRAGEN output equivalency across AWS F1 and F2 instance families, we evaluated the output for five samples processed on each instance type and compared outputs at three levels: pipeline metrics, command provenance, and variant calls. For like-for-like runs, we expect identical results across instances. In practice, every value and variant should match exactly, with differences limited to non-deterministic metadata such as timestamps, file paths, or sample labels.
Command Provenance: To verify that the pipelines were invoked identically, we extracted the DRAGEN command-lines from the VCF headers and compared them using bash tools. The commands and flags matched exactly between F1 and F2 runs, ensuring the analysis was conducted with the same configuration and parameters.
Metrics Comparison: We compared DRAGEN-generated metrics using standard bash utilities (e.g., diff) across the following files: gc_metrics, mapping_metrics, roh_metrics, sv_metrics, and vc_metrics. Across all five samples, the metrics were identical between F1 and F2 outputs except for the values we expect to vary (e.g., names, date/timestamp). This confirms that alignment, variant calling, and summary statistics are reproducible across both instance types.
VCF Concordance: We evaluated variant-level concordance using bcftools isec, which performs set operations on VCF files by treating each file as a set of variant records. It computes intersections (variants present in both files) and differences (variants unique to one file), making it a practical way to quantify concordance between outputs. In our workflow, we ran bcftools isec on paired VCFs (F1 vs F2) for each sample. The tool writes out separate files corresponding to the following:
- Variants unique to the first VCF (e.g., 0000.vcf)
- Variants unique to the second VCF (e.g., 0001.vcf)
- Variants common to both VCFs (the intersection, e.g., 0002.vcf, and 0003.vcf)
We then counted variants in each output to measure overlap and differences, excluding header lines that begin with # or ##. Under full equivalence, the unique outputs should be empty, and the intersection files should contain all variants, so intersection record counts match the originals.
# Run bcftools isec
$> bcftools isec -p results sample1_f1.vcf.gz sample1_f2.vcf.gz
# Count variant records (exclude header lines)
$> grep -c -v "^#" results/*.vcf
0000.vcf:0
0001.vcf:0
0002.vcf:299131
0003.vcf:299131Across all five samples, bcftools isec showed that the intersection contained all variants, and the unique sets were empty, indicating no variant-level differences between F1 and F2 outputs. In other words, the VCFs were fully concordant.
In conclusion, under identical DRAGEN configurations, Amazon EC2 F1 and F2 instances produce equivalent outputs at both the metrics and variant levels. Metrics differed only in expected, non-deterministic metadata (timestamps and sample labels), and bcftools isec showed that variant intersections contained all calls with no unique differences. These results support running DRAGEN genomic pipelines interchangeably on F1 and F2.
The impact: accelerating genomic discovery
The data suggest that successful migration includes advantages that extend beyond simpler infrastructure upgrades. The immediate benefits include establishing a clear, validated migration path from F1 to F2 instances, ensuring business continuity and achieving significant cost and CO2 emissions optimization for large-scale genomic workloads.
Conclusion
The results demonstrate how cloud infrastructure evolution can drive efficiency improvements in genomic research capabilities. The 60% performance improvement, 70% cost reduction, as well as improved sustainability achieved through F2 instance migration provides compelling data that may be useful for organizations looking to optimize their genomic processing workloads.
For genomics teams currently using F1 instances, this validation study offers confidence in migrating to F2 instances while maintaining research integrity and achieving substantial operational benefits.
To learn more about AWS F2 instances for genomics workloads, visit Genomics on AWS. For DRAGEN implementation guidance, explore the Illumina DRAGEN product information page.
[1] DRAGEN® is a registered trademark of Illumina Inc.