AWS HPC Blog
Optimize Nextflow Workflows on AWS Batch with Mountpoint for Amazon S3
This post was contributed by Michael Mueller (AWS) and Simon Murray (Genomics England)
Are you running genomic workflows with Nextflow on AWS Batch and experiencing bottlenecks when staging large reference files? In this post, we will show you how to optimize your workflow performance by leveraging Mountpoint for Amazon S3 to stream reference data directly into your Nextflow processes, eliminating the need to stage large static files repeatedly.
Background
When running Nextflow workflows on AWS Batch, input files typically need to be staged from Amazon Simple Storage Service (Amazon S3) to Amazon Elastic Block Store (Amazon EBS). For large reference files, this staging process can create significant bottlenecks, especially when the same files are copied repeatedly for different process executions.
Mountpoint for Amazon S3: a better approach
Mountpoint for Amazon S3 is a file client that provides a POSIX-like interface to Amazon S3 object storage. It’s specifically optimized for read access to large objects, making it ideal for genomic workflows that require access to large reference files. With Mountpoint, you can mount an S3 bucket as a local file system and stream data directly into your processes without copying the entire file to local storage first.
Understanding Nextflow data handling
In Nextflow, there are two ways to reference input files:
- Dataflow Channels: The recommended approach for files that are passed between processes and represent intermediate results that can create dependencies between workflow tasks that Nextflow manages when orchestrating the workflow execution.
- Direct Path References: Suitable for static reference files that do not change between runs and are not passed between processes and therefore do not impact orchestration of workflow tasks.
How Genomics England leverages Mountpoint for Amazon S3 to accelerate their variant annotation pipeline
Genomics England employs a Nextflow workflow that runs the Variant Effect Predictor tool, which requires over 500 GB of reference data. Initially, the team considered using Nextflow’s built in S3 integration. However, this approach quickly proved impractical due to the lengthy staging times for such large datasets. To overcome this challenge, Genomics England adopted Mountpoint for Amazon S3 for the reference data, eliminating the need for time-consuming file staging. This optimization significantly accelerated their variant annotation pipeline.
Solution architecture
Figure 1 shows how reference data stored in an Amazon S3 bucket is accessed from a Nextflow process running in a Docker container using Mountpoint for Amazon S3. Other input data is staged to the Amazon EBS volume attached to the Amazon Elastic Compute Cloud (Amazon EC2) instance.
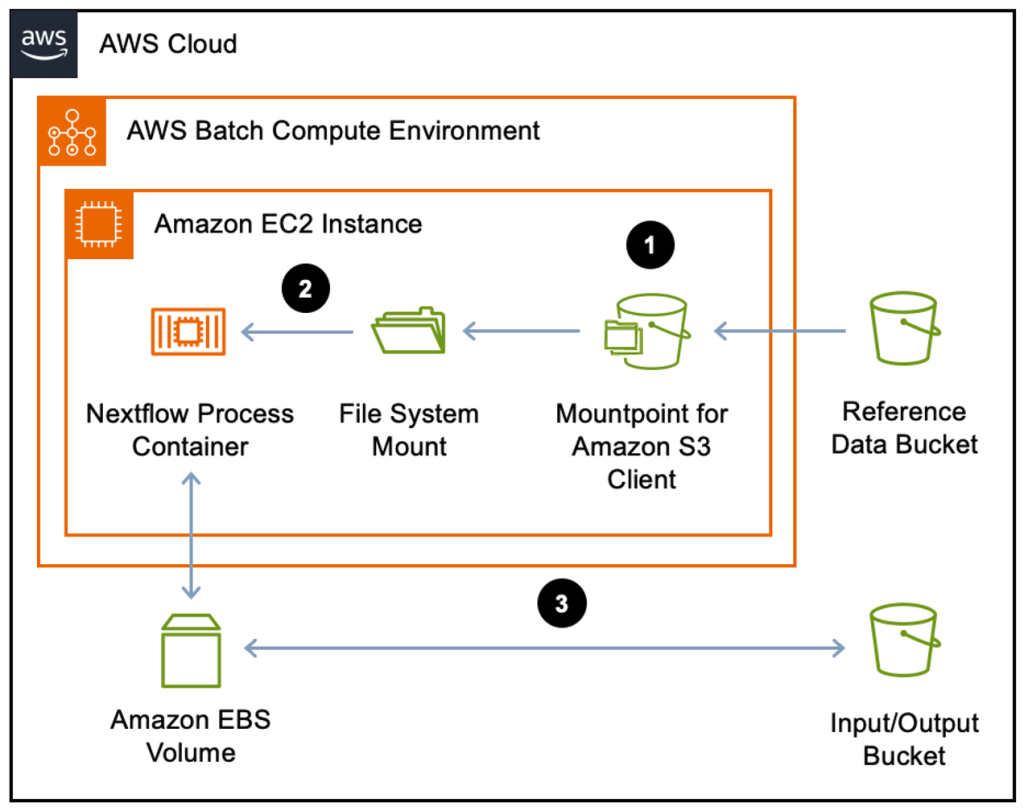
Figure 1: AWS Batch using Mountpoint for Amazon S3 architecture. Reference data is streamed into the Nextflow process container via the Mountpoint for Amazon S3 client that presents the Amazon S3 bucket as a directory on the local file system (1). The mount path is mapped into the Docker container as a volume (2). Other input data is staged to the Amazon EBS volume attached to the Amazon EC2 instance through Nextflow’s standard Amazon S3 integration (3).
Here’s how to set up your environment to use Mountpoint for Amazon S3 in your Nextflow workflows.
Prerequisites
- An AWS account with appropriate permissions
- AWS Batch environment configured
- Nextflow (community edition) installed
(Nextflow 24.10.4 with nf-amazon plugin version 2.9.0 was used for this blog post) - Access to reference data in an S3 bucket
Walkthrough
1. Add Mountpoint client installation to EC2 launch template
You should already have a launch template associated with your AWS Batch compute environment. In the User Data section add the bash commands that will install the Mountpoint client and mount your S3 bucket:
Please note Mountpoint requires the name of an S3 bucket not the S3 URI.
Running Docker containers as non-root users is considered a security best practice as it reduces the potential impact of container breakouts and follows the principle of least privilege. By default, users other than the user who ran the mount-s3 command cannot access your mounted directory, even if the permissions and ownership settings would allow it. This is true even for the root user and is a limitation of the FUSE system Mountpoint uses to create a file system.
When running Docker containers as non-root users, modify your Mountpoint command in the EC2 launch template to allow non-root users access to the mount:
To use this flag, you may need to first configure FUSE by adding the line user_allow_other to the /etc/fuse.conf file. Even with this flag enabled, Mountpoint still respects the permissions and ownership configured with other flags.
2. Configure Nextflow volume
In the AWS Batch configuration of your Nextflow pipeline, add the Mountpoint for Amazon S3 directory as a volume:
3. Pass reference file path to Nextflow process
To stream data directly from Amazon S3 without staging, the path to the Mountpoint for Amazon S3 directory must be passed into the Nextflow process using the val input directive. (Note that using the path input directive with a Mountpoint path will result in an error during data staging.) However, continue to pass non-static data using the path directive.
Mountpoint paths passed as val can be referenced directly in the script block:
Build a custom AMI to streamline the setup
To optimize your infrastructure deployment you can create a custom Amazon Machine Image (AMI) with pre-installed Mountpoint for Amazon S3. This approach eliminates the need for repetitive downloading of the Mountpoint client during each EC2 instance launch. By packaging Mountpoint into your custom AMI, you can ensure faster instance startup times and standardize your deployment configuration across your compute environment.
Create your custom AMI from the launch template through the AWS Management Console:
- Navigate to the EC2 Dashboard
- Locate the “Launch Templates” option in the left navigation pane
- Select your Mountpoint launch template
- Use the “Actions” dropdown
- Choose “Launch instance from template.”
Once your instance is running with Mountpoint for Amazon S3 properly installed and configured, create your custom AMI:
- Select the instance in the EC2 console
- Choose “Actions”
- Select “Image and templates”
- Choose “Create image” to create your AMI
Now, update your launch template to remove the Mountpoint setup from the User Data section and specify your custom AMI in the Application and OS Images section.
Conclusion
In this post, we showed how to optimize Nextflow workflows on AWS Batch by using Mountpoint for Amazon S3 to stream reference data. This approach eliminates file staging bottlenecks for large static reference files in your genomic pipelines. To get started and learn more about mounting S3 buckets as a local file system, visit the Mountpoint for Amazon S3 documentation and Github repository.