AWS HPC Blog
Scalable Cryo-EM on AWS Parallel Computing Service (PCS)
 Cryogenic electron microscopy (Cryo-EM) allows drug discovery researchers to determine the three-dimensional structures of molecules critical to drug discovery. As Cryo-EM adoption has increased, scientists and IT owners have looked for ways to efficiently process the terabytes (and terabytes) of data generated each day by these microscopes. These processing pipelines need scalable, heterogeneous compute combined with both fast and cost-effective storage.
Cryogenic electron microscopy (Cryo-EM) allows drug discovery researchers to determine the three-dimensional structures of molecules critical to drug discovery. As Cryo-EM adoption has increased, scientists and IT owners have looked for ways to efficiently process the terabytes (and terabytes) of data generated each day by these microscopes. These processing pipelines need scalable, heterogeneous compute combined with both fast and cost-effective storage.
AWS Parallel Computing Service (PCS) is a managed service for deploying and managing HPC clusters in the cloud. Using PCS for Cryo-EM can reduce the undifferentiated heavy lifting of building and managing HPC infrastructure while maintaining a consistent user experience for structural biologists, who can get to work on their research quickly.
In this post, we’re presenting a recommended reference architecture you can use for Cryo-EM on PCS, and we’ll provide a specific example with CryoSPARC: a popular application. We’ll also show ChimeraX for visualization, and cover some best-practices for running Cryo-EM in the cloud generally.
Architecture overview
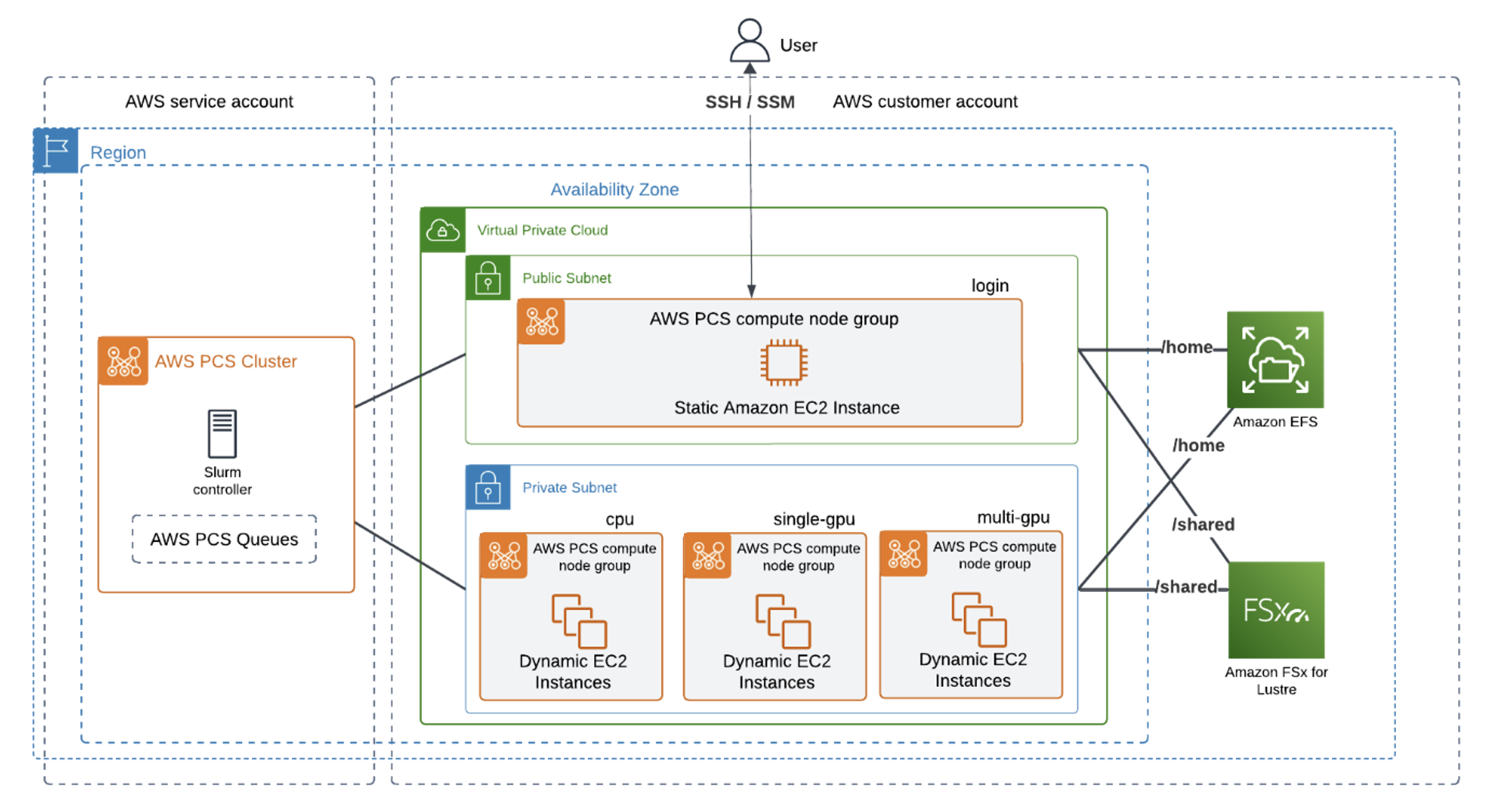
Figure 1 – Architecture overview of CryoSPARC on AWS. The Slurm controller is deployed in an AWS service account, and the compute and storage resources are deployed in the user AWS account. FSx for Lustre and Amazon Elastic File Store (EFS) are both mounted to the cluster.
Setup & pre-requisites
In addition to the prerequisites listed in the PCS documentation, you’ll need a CryoSPARC license. It’s possible to follow this guide through PCS cluster creation without a license, but ultimately you’ll need one to install the software and run a test job. To obtain a license, contact Structura Biotechnology.
Create a cluster with shared storage
The HPC Recipes Library is a public GitHub repository that shares templates created by our engineering and architecture teams for deploying HPC infrastructure on the cloud without steep learning curves. To create a PCS cluster with the right shared storage for this example, you can use the PCS guidance for a one-click deployment, which uses AWS CloudFormation to launch an entire cluster, quickly.
When CloudFormation opens, you’ll have the option to provide an SSH key for accessing the cluster login node. Leave all other fields as-is, and choose Create. This will create the required networking prerequisites, a cluster with a Login Node group, a single demo Compute Node group, an EFS file system for /home, and a Lustre file system for /shared.
Once it’s ready, you should see the following stacks in your CloudFormation console. Figure 2 shows a screenshot from CloudFormation, including a short description of what’s deployed in each stack.

Figure 2: CloudFormation stacks created by hpc recipes template.
Alternatively, if you want to create a PCS cluster manually or using existing resources in your account, just follow the steps in the PCS User Guide to set up these resources.
Adjust FSx for Lustre file system throughput
In your CloudFormation console, click the View Nested radio slider to see the various stacks that have been created for you from the template you deployed. Find the stack that starts with the name get-started-cfn-FSxLStorage and click on it. When the stack information displays on the right-hand side of your console, click on the Outputs tab and note the value of FSxLustreFilesystemId to use in a moment.
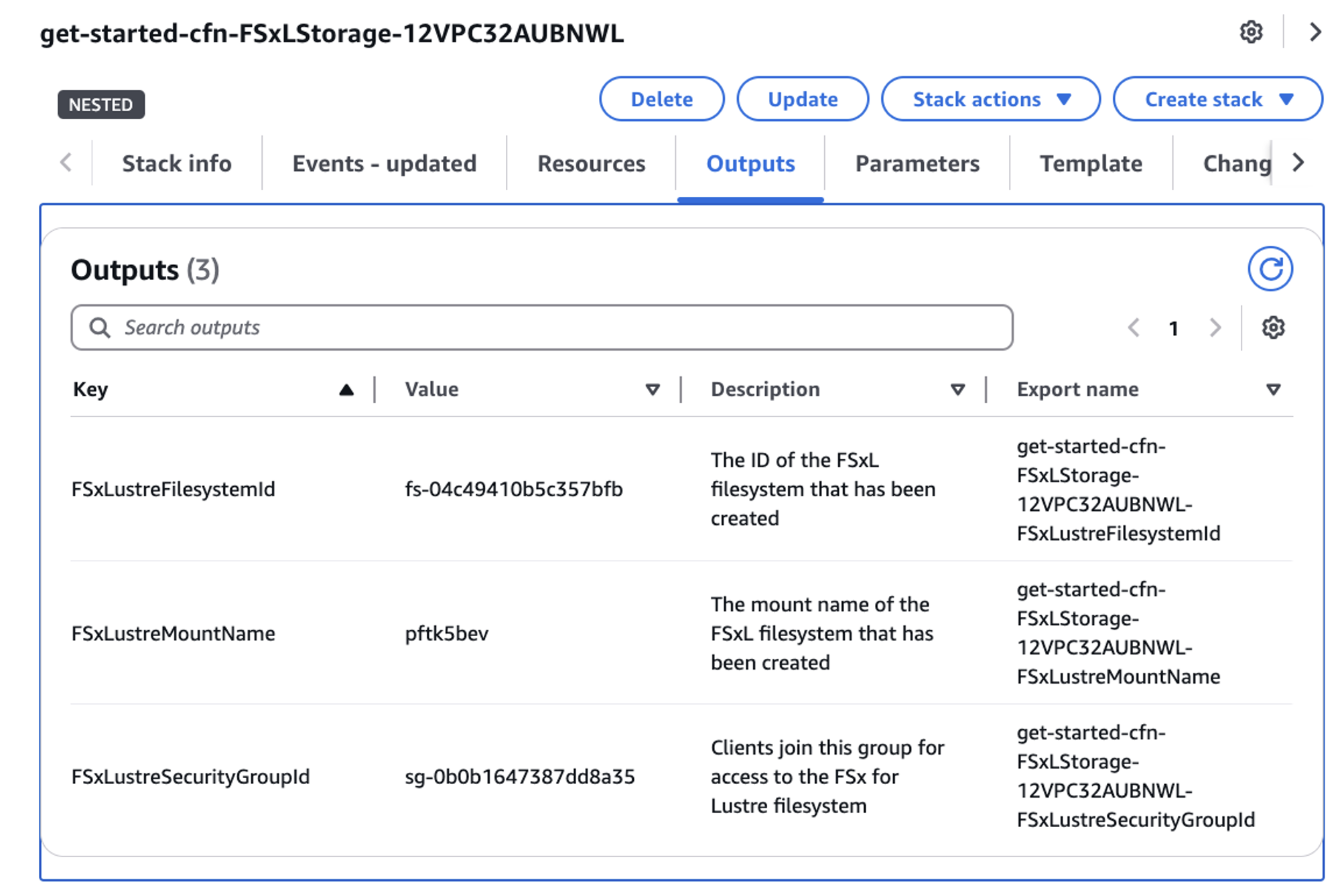
Figure 3: FSx for Lustre CloudFormation stack created by hpc recipes template.
We will need to update the throughput per unit of storage to 250 MB/s/TiB for our FSx for Lustre system to achieve a successful install of CryoSPARC. This can take up to 20 minutes so let’s run the command now to give the file system time to update in the background while we setup the rest of our cluster.
aws fsx update-file-system \
--file-system-id $FSX-LUSTRE-ID \
--lustre-configuration PerUnitStorageThroughput=250
Create additional node groups and queues
Once our initial cluster creation is complete, we can create some compute node groups and queues. An AWS PCS compute node group is a logical collection of nodes (instances, as we call them) from the Amazon Elastic Compute Cloud (Amazon EC2). These are the ephemeral machines where your will be executed. An AWS PCS queue is a lightweight abstraction over the scheduler’s native implementation of a work queue. Jobs are submitted to queue, which is mapped to one or more compute node groups. In CryoSPARC, lanes are equivalent to PCS queues.
We will create three new compute node groups; compute-cpu (with c5a.8xlarge instances), compute-single-gpu (g6.4xlarge), and compute-multi-gpu (g6.48xlarge) and map those compute node groups to respective queues. These instance types were chosen based on our own internal testing. For a detailed explanation of scalability for individual tasks within the processing pipeline, you can see the CryoSPARC performance benchmarks that explain these selection decisions.
You can create these node groups from the PCS console, but today, we’ll show you how to do it with the AWS CLI. Execute this command to get the AMI ID, Instance Profile, and Launch Template ID of the compute-1 PCS Compute Node Group and save the output. We’ll use this in the next set of commands to create additional compute node groups:
aws pcs get-compute-node-group \
--cluster-identifier $PCS_CLUSTER_NAME \
--compute-node-group-identifier compute-1
Run these, and save the compute node group name and id from the outputs for each command. We’ll use this to map these node groups to queues:
aws pcs create-compute-node-group \
--compute-node-group-name compute-cpu \
--cluster-identifier $PCS_CLUSTER_NAME \
--region $REGION \
--subnet-ids $PRIVATE_SUBNET_ID \
--custom-launch-template id=$COMPUTE_LT_ID,version='1' \
--ami-id $AMI_ID \
--iam-instance-profile $INSTANCE_PROFILE_ARN \
--scaling-config minInstanceCount=0,maxInstanceCount=2 \
--instance-configs instanceType=c5a.8xlarge
aws pcs create-compute-node-group \
--compute-node-group-name compute-single-gpu \
--cluster-identifier $PCS_CLUSTER_NAME \
--region $REGION \
--subnet-ids $PRIVATE_SUBNET_ID \
--custom-launch-template id=$COMPUTE_LT_ID,version='1' \
--ami-id $AMI_ID \
--iam-instance-profile $INSTANCE_PROFILE_ARN \
--scaling-config minInstanceCount=0,maxInstanceCount=2 \
--instance-configs instanceType=g6.4xlarge
aws pcs create-compute-node-group \
--compute-node-group-name compute-multi-gpu \
--cluster-identifier $PCS_CLUSTER_NAME \
--region $REGION \
--subnet-ids $PRIVATE_SUBNET_ID \
--custom-launch-template id=$COMPUTE_LT_ID,version='1' \
--ami-id $AMI_ID \
--iam-instance-profile $INSTANCE_PROFILE_ARN \
--scaling-config minInstanceCount=0,maxInstanceCount=2 \
--instance-configs instanceType=g6.48xlarge
Check the status of the node group creation by running this command:
aws pcs get-compute-node-group --region $region \
--cluster-identifier $cluster-name \
--compute-node-group-identifier $node-group-name
When the status for each of the three node groups is ACTIVE, you can move forward to creating your queues. Each queue is mapped to one or more node groups, which are responsible for supplying ephemeral instances for the jobs arriving in the queue. For this cluster, we map each queue to a single node group.
Note that the $NODE_GROUP_ID is not same as the node group name.
aws pcs create-queue \
--queue-name cpu-queue \
--cluster-identifier $PCS_CLUSTER_NAME \
--compute-node-group-configurations
computeNodeGroupId=$COMPUTE_CPU_NODE_GROUP_ID
aws pcs create-queue \
--queue-name single-gpu-queue \
--cluster-identifier $PCS_CLUSTER_NAME \
--compute-node-group-configurations computeNodeGroupId=$COMPUTE_SINGLE_GPU_NODE_GROUP_ID
aws pcs create-queue \
--queue-name multi-gpu-queue \
--cluster-identifier $PCS_CLUSTER_NAME \
--compute-node-group-configurations computeNodeGroupId=$COMPUTE_MULTI_GPU_NODE_GROUP_ID
Next, confirm your queue created successfully:
aws pcs get-queue --region $REGION \
--cluster-identifier $PCS_CLUSTER_NAME \
--queue-identifier $PCS_QUEUE_NAME
Once the status returns ACTIVE, your queue creation is complete. Now we can log into a cluster login node and install CryoSPARC.
Open the Amazon EC2 console and navigate to Instances. Search for aws:pcs:compute-node-group-id = <LOGIN_COMPUTE_NODE_GROUP_ID> in the search bar making sure to replace <LOGIN_COMPUTE_NODE_GROUP_ID> with the ID of your login compute node group and hit enter. Select this instance, and then select Connect. On the next page, choose Session Manager and then Connect. This will open a terminal session in a browser tab (this is a great feature of Session Manager). In the terminal, change the user to ec2-user, which is the user in the cluster that has privileges with Slurm to submit and manage jobs.
sudo su - ec2-userOnce you’re connected to your cluster’s login node, you should now see the additional Slurm partitions by running this command:
sinfoWhich should show:
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
demo up infinite 4 idle~ compute-1-[1-4]
single-GPU up infinite 4 idle~ single-GPU-[1-4]
CPU up infinite 4 idle~ CPU-[1-4]
multi-GPU up infinite 4 idle~ multi-GPU-[1-4]Now that you’re logged onto the cluster, we’re ready to install CryoSPARC and download a test dataset.
Installing CryoSPARC and downloading a test dataset
To simplify CryoSPARC installation and setup, we’ve provided a shell script that installs the application on the shared file system and registers lanes based on the cluster queue names. To access this, generate a key pair and SSH to your login node. When connected to your login node, download the script and make it an executable:
wget https://raw.githubusercontent.com/aws-samples/cryoem-on-aws-parallel-cluster/refs/heads/main/parallel-computing-service/pcs-cryosparc-post-install.shRun the script, pointing to the shared file system for installation. Replace the $LICENSE_ID with the license your CryoSPARC license. Note that this can take up to an hour.
chmod +x pcs-cryosparc-post-install.sh
sudo ./pcs-cryosparc-post-install.sh $LICENSE_ID /shared/cryosparc /shared/cuda 11.8.0 11.8.0_520.61.05 /shared
Once installation is complete, start the CryoSPARC server:
/shared/cryosparc/cryosparc_master/bin/cryosparcm startIf you reboot your login node, you will need to run the start command for the CryoSPARC server again. This command can be added to the EC2 user data section of your launch template to automate this process. You can find more information on working with Amazon EC2 user data in the PCS User Guide.
After the server has successfully started with the confirmation message CryoSPARC master started, create a new user:
cryosparcm createuser \
--email "<youremail@email.com>" \
--password "<yourpassword>" \
--username "<yourusername>" \
--firstname "yourname>" \
--lastname "<yourlastname>"
Once done, exit out of the login node.
Access the CryoSparc UI
Next, set up an SSH tunnel using your previously generated EC2 key pair to our (now) CryoSPARC login node, which will let us connect to the CryoSPARC web interface:
ssh -i /path/to/key/key-name -N -f -L \ localhost:45000:localhost:45000 ec2-user@publicIPofyourinstanceAfter executing this successfully, you can visit http://localhost:45000 in your web browser which should now tunnel you to the login screen for CryoSPARC.
Figure 4: From the web browser login page, access CryoSPARC using the newly create user credentials.
Run a test job
You can create a data folder for your test data in the /shared directory and download the test data set using these commands:
mkdir /shared/data
cd /shared/data
/shared/cryosparc/cryosparc_master/bin/cryosparcm downloadtest
tar -xf empiar_10025_subset.tar
This step will take a few minutes to complete.
For this test, we’re downloading the dataset directly to the Lustre file system. In a production environment, we recommend storing datasets in Amazon Simple Storage Service (Amazon S3), and using a Data Repository Association (DRA) between Amazon S3 and the Amazon Fsx for Lustre file system. Since a single Cryo-EM sample can be tens of terabytes in size - and organizations regularly store petabytes of microscopy data - using Amazon S3 with FSx for Lustre like this can save you a lot of money. To set up a DRA, see the FSx for Lustre documentation.
Once the test dataset is downloaded, follow the steps in the Get Started with CryoSPARC Tutorial to run an Import Movies job. Note that when selecting a queue for the job, you will see the queues from your Slurm cluster. Select the compute-cpu lane for the Import Movies job:
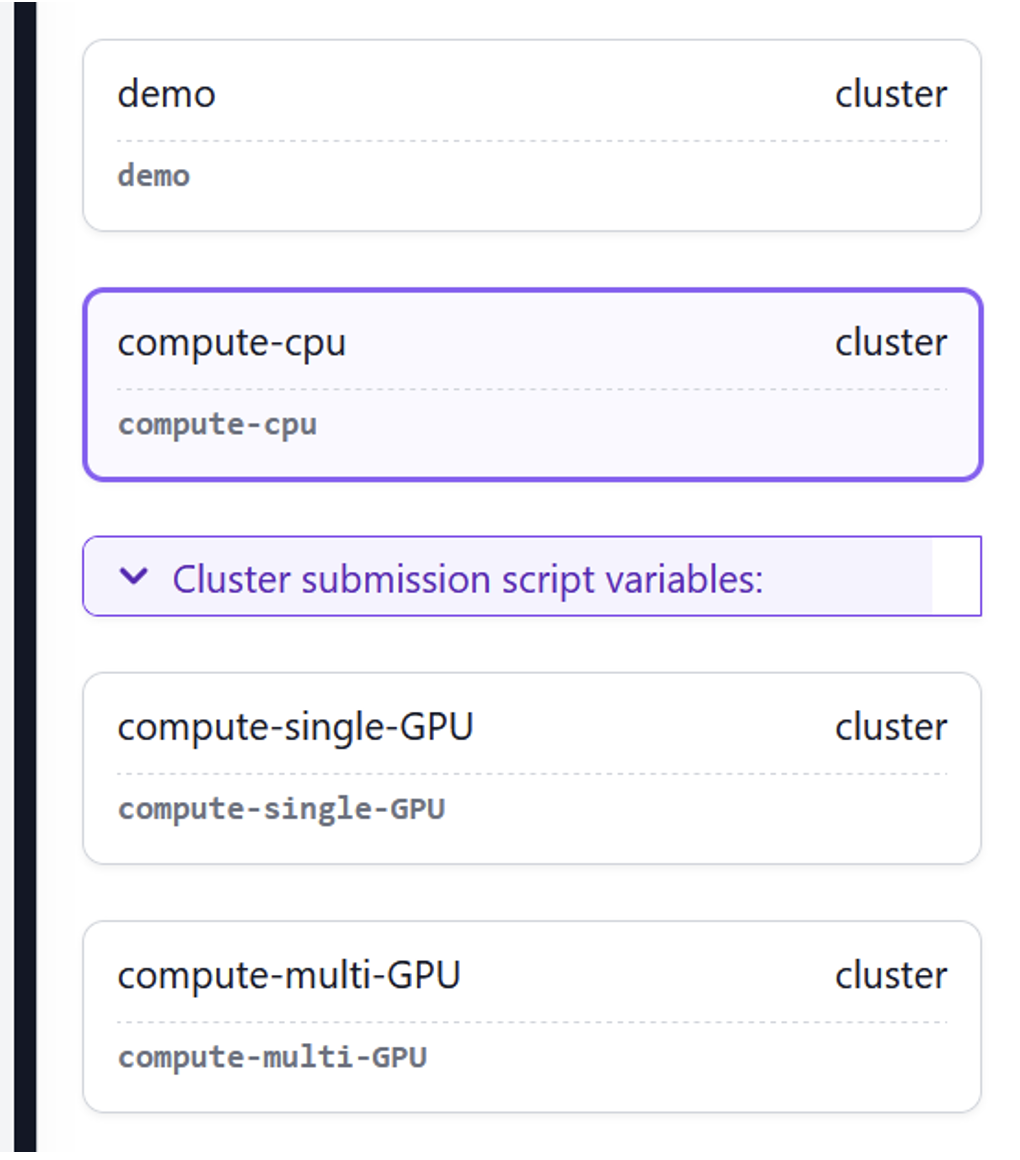
Figure 5: CryoSPARC configured lanes with the same names as PCS queues as part of the installation. Choose compute-cpu for the Import Movies job.
Now, run the job. Under Event Log in the CryoSPARC UI, you should see a Slurm submission like this:
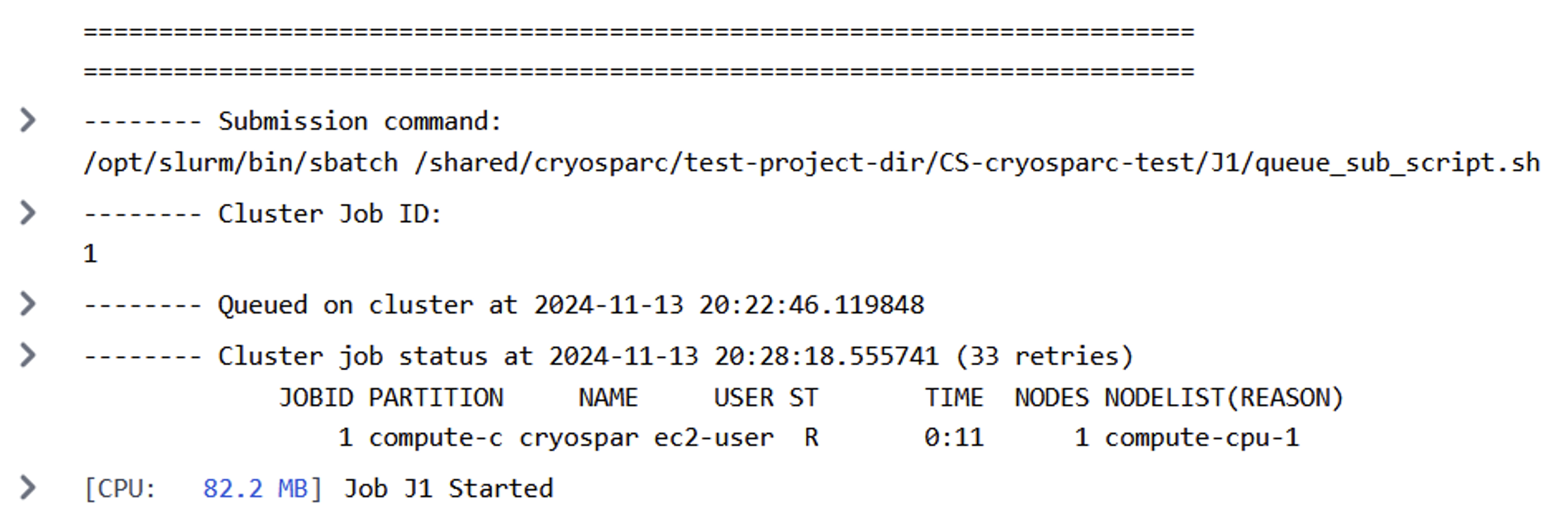
Figure 6: Successful CryoSPARC job submission.
Back in the terminal, from the login node, you can run the squeue command to see the job executing on the cluster:
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
1 compute-c cryospar ec2-user CF 1:02 1 compute-cpu-1
Run sinfo to see the single node being allocated for the job:
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
demo up infinite 4 idle~ compute-1-[1-4]
compute-cpu up infinite 1 mix# compute-cpu-1
compute-cpu up infinite 3 idle~ compute-cpu-[2-4]
compute-single-GPU up infinite 4 idle~ compute-single-gpu-[1-4]
compute-multi-GPU up infinite 4 idle~ compute-multi-gpu-[1-4]
This node is a single instance provisioned in your AWS account by EC2. You can check the EC2 console to see it. The job should successfully complete with a few minutes. If you don’t submit any further jobs to the queue, you’ll see that instance dynamically terminated a few minutes after your last job completes.
Visualization and next steps
We can install additional applications - like visualization packages - on the cluster shared storage.
ChimeraX is a commonly-used visualization application for structural biologists. We don’t cover it in this post, but you can run this on the cluster by configuring Amazon DCV on the login node. DCV provides low-latency, high-resolution remote visualization between your desktop and the cloud, removing the need for time-intensive and costly data movement between your site and cloud.
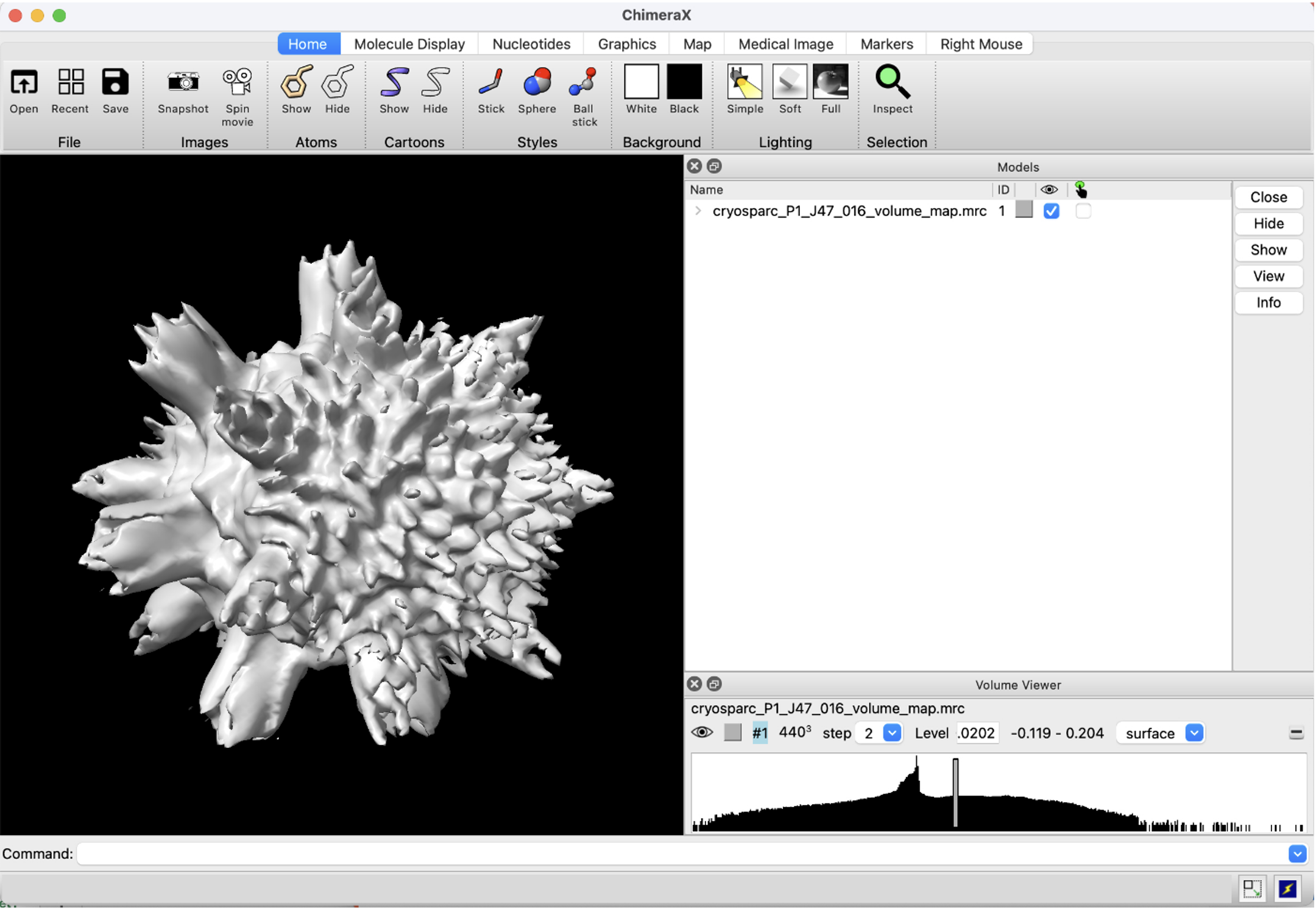
Figure 7: Screenshot of results from EMPIAR 10288 Sample, run through CryoSPARC, and visualized using ChimeraX.
Tear down
Using the AWS CLI, you can tear down everything built in this post by first deleting the cpu-queue, single-gpu-queue, and multi-gpu-queue using this command:
aws pcs delete-queue --cluster-identifier <pcs_cluster_name> --queue-identifier <pcs_queue_name>Next, delete the compute-cpu, compute-single-gpu, and compute-multi-gpu compute node groups using this command:
aws pcs delete-queue --cluster-identifier <pcs_cluster_name> --compute-node-group-identifier <pcs_compute_node_group_name>Finally, delete the PCS cluster and all resources created with it by deleting your CloudFormation template using this command:
aws cloudformation delete-stack --stack-name <pcs_cloudformation_stack name>Conclusion
AWS Parallel Computing Service provides a powerful and scalable solution for running Cryo EM in the cloud, enabling researchers to unlock new scientific discovery.
With scalable, on-demand computing on AWS, you can meet the demands of your scientists as their ideas – and ambitions - grow. You can configure AWS PCS with heterogeneous compute architectures and keep it up to date with the latest instance types as they become available. High-resolution, low-latency visualization integrated into PCS using Amazon DCV can enable scientists to execute their full Cryo-EM workflows directly from their desktops.
Customers choose AWS for their Cryo-EM for these reasons, because they result in scalability, flexibility, end efficiency for their researchers.
This guide walked through a single example of how to run a Cryo-EM job on PCS. Structural biologists often use multiple applications when processing a single sample, and share datasets across research groups within an organization. Amazon Professional Services and our Amazon Partner Network (APN) members like CloverTex can help scale out this initial system to meet your organization’s needs. To learn more, contact your AWS account team or reach out to find us at ask-hpc@amazon.com.