AWS Big Data Blog
Unlock the power of Apache Iceberg v3 deletion vectors on Amazon EMR
As modern data architectures expand, Apache Iceberg has become a widely popular open table format, providing ACID transactions, time travel, and schema evolution. In table format v2, Iceberg introduced merge-on-read, improving delete and update handling through positional delete files. These files improve write performance but can slow down reads when not compacted, since Iceberg must merge them during query execution to return the latest snapshot. Iceberg v3 enhances merge performance during reads by replacing positional delete files with deletion vectors for handling row-level deletes in Merge-on-Read (MoR) tables. This change deprecates the use of positional delete files in v3, which marked specific row positions as deleted, in favor of the more efficient deletion vectors.
In this post, we compare and evaluate the performance of the new binary deletion vectors in Iceberg v3 with respect to traditional position delete files of Iceberg v2 using Amazon EMR version 7.10.0 with Apache Spark 3.5.5. We provide insights into the practical impacts of these advanced row-level delete mechanisms on data management efficiency and performance.
Understanding binary deletion vectors and Puffin files
Binary deletion vectors stored in Puffin files use compressed bitmaps to efficiently represent which rows have been deleted within a data file. In contrast, previous Iceberg versions (v2) relied on positional delete files—Parquet files that enumerated rows to delete by file and position. This older approach resulted in many small delete files, which placed a heavy burden on query engines due to numerous file reads and costly in-memory conversions. Puffin files reduce this overhead by compactly encoding deletions, improving query performance and resource utilization.
Iceberg v3 improves this in the following aspects:
- Reduced I/O – Fewer small delete files lower metadata overhead by introducing deletion vectors—compressed bitmaps that efficiently represent deleted rows. These vectors are stored persistently in Puffin files, a compact binary format optimized for low-latency access.
- Query performance – Bitmap-based deletion vectors enable faster scan filtering by allowing multiple vectors to be stored in a single Puffin file. This reduces metadata and file count overhead while preserving file-level granularity for efficient reads. The design supports continuous merging of deletion vectors, promoting ongoing compaction that maintains stable query performance and reduces fragmentation over time. It removes the trade-off between partition-level and file-level delete granularity seen in v2, enabling consistently fast reads even in heavy-update scenarios.
- Storage efficiency – Iceberg v3 uses a compressed binary format instead of verbose Parquet positioning. Engines maintain a single deletion vector per data file at write time, enabling better compaction and consistent query performance.
Solution overview
To explore the performance characteristics of delete operations in Iceberg v2 and v3, we use PySpark to run our comparison tests focusing on delete operation runtime and delete file size. This implementation helps us effectively benchmark and compare the deletion mechanisms between Iceberg v2’s position-delete files using Parquet and v3’s newer Puffin-based deletion vectors.
Our solution demonstrates how to configure Spark with the AWS Glue Data Catalog and Iceberg, create tables, and run delete operations programmatically. We first create Iceberg tables with format versions 2 and 3, insert 10,000 rows, then perform delete operations on a range of record IDs. We also perform table compaction and then measure delete operation runtime and size and count of associated delete files.
In Iceberg v3, deleting rows introduces binary deletion vectors stored in Puffin files (compact binary sidecar files). These allow more efficient query planning and faster read performance by consolidating deletes and avoiding large numbers of small files.
For this test, the Spark job was submitted by SSH’ing into the EMR cluster and using spark-submit directly from the shell, with the required Iceberg JAR file being referenced directly from the Amazon Simple Storage Service (Amazon S3) bucket in the submission command. When running the job, make sure you provide your S3 bucket name. See the following code:
Prerequisites
To follow along with this post, you must have the following prerequisites:
- Amazon EMR on Amazon EC2 with version 7.10.0 integrated with the Glue Data Catalog, which includes Spark 3.5.5.
- The Iceberg 1.9.2 JAR file from the official Iceberg documentation, which includes important deletion vector improvements such as v2 to v3 rewrites and dangling deletion vector detection. Optionally, you can use the default Iceberg 1.8.1-amzn-0 bundled with Amazon EMR 7.10 if these Iceberg 1.9.x improvements are not required.
- An S3 bucket to store Iceberg data.
- An AWS Identity and Access management (IAM) role for Amazon EMR configured with the necessary permissions.
The upcoming Amazon EMR 7.11 will ship with Iceberg 1.9.1-amzn-1, which includes deletion vector improvements such as v2 to v3 rewrites and dangling deletion vector detection. This means you no longer need to manually download or upload the Iceberg JAR file, because it will be included and managed natively by Amazon EMR.
Code walkthrough
The following PySpark script demonstrates how to create, write, compact, and delete records in Iceberg tables with two different format versions (v2 and v3) using the Glue Data Catalog as the metastore. The main goal is to compare both write and read performance, along with storage characteristics (delete file format and size) between Iceberg format versions 2 and 3.
The code performs the following functions:
- Creates a SparkSession configured to use Iceberg with Glue Data Catalog integration.
- Creates a synthetic dataset simulating user records:
- Uses a fixed random seed (42) to provide consistent data generation
- Creates identical datasets for both v2 and v3 tables for fair comparison
- Defines the function test_read_performance(table_name) to perform the following actions:
- Measure full table scan performance
- Measure filtered read performance (with WHERE clause)
- Track record counts for both operations
- Defines the function test_iceberg_table(version, test_df) to perform the following actions:
- Create or use an Iceberg table for the specified format version
- Append data to the Iceberg table
- Trigger Iceberg’s data compaction using a system procedure
- Delete rows with IDs between 1000–1099
- Collect statistics about inserted data files and delete-related files
- Measure and record read performance metrics
- Track operation timing for inserts, deletes, and reads
- Defines a function to print a comprehensive comparative report including the following information:
- Delete operation performance
- Read performance (both full table and filtered)
- Delete file characteristics (formats, counts, sizes)
- Performance improvements as percentages
- Storage efficiency metrics
- Orchestrate the main execution flow:
- Create a single dataset to ensure identical data for both versions
- Clean up existing tables for fresh testing
- Run tests for Iceberg format version 2 and version 3
- Output a detailed comparison report
- Handle exceptions and shut down the Spark session
See the following code:
Results summary
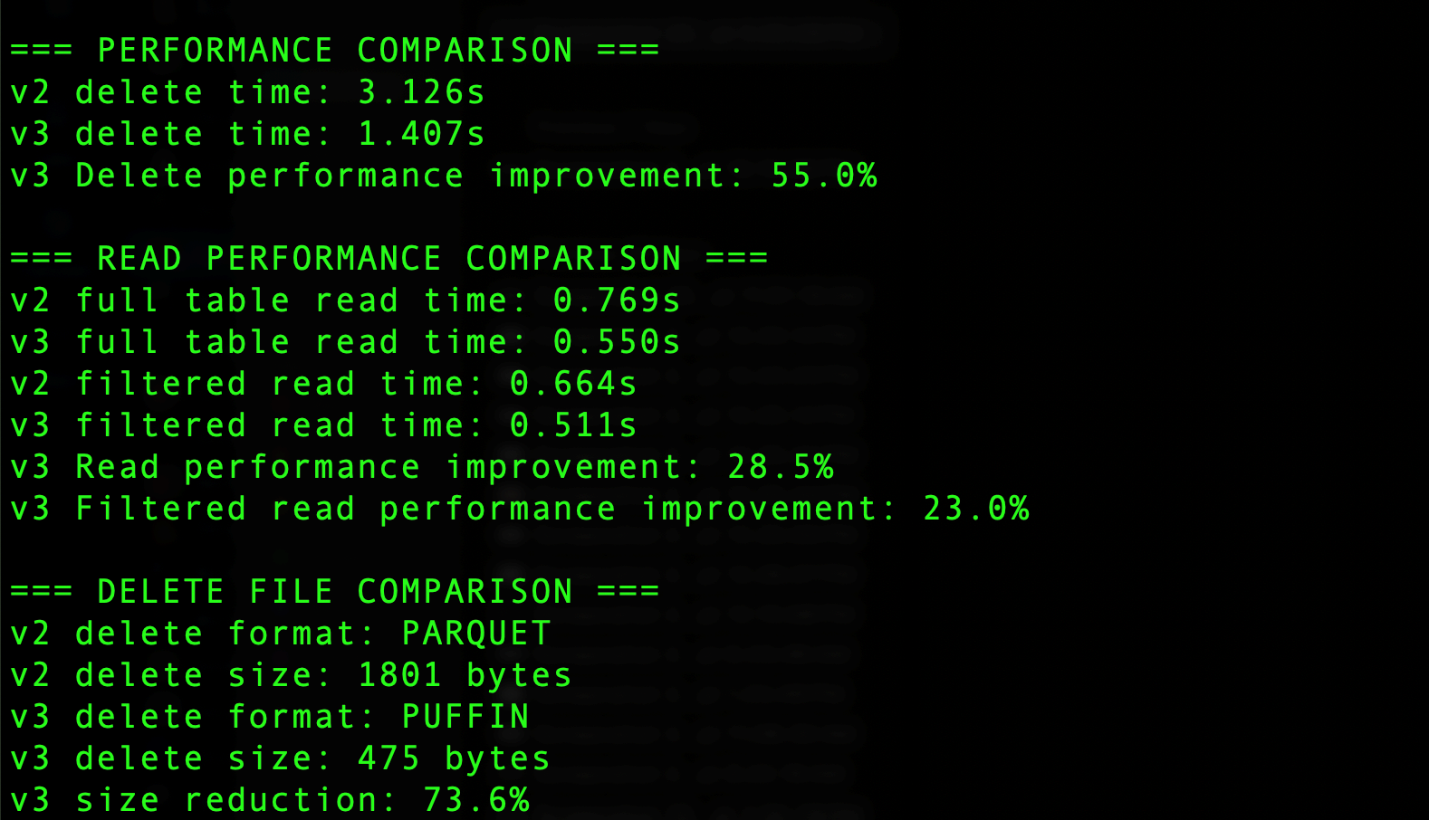
The output generated by the code includes the results summary section that shows several key comparisons, as shown in the following screenshot. For delete operations, Iceberg v3 uses the Puffin file format compared to Parquet in v2, resulting in significant improvements. The delete operation time decreased from 3.126 seconds in v2 to 1.407 seconds in v3, achieving a 55.0% performance improvement. Additionally, the delete file size was reduced from 1801 bytes using Parquet in v2 to 475 bytes using Puffin in v3, representing a 73.6% reduction in storage overhead. Read operations also saw notable improvements, with full table reads 28.5% faster and filtered reads 23% faster in v3. These improvements demonstrate the efficiency gains from v3’s implementation of binary deletion vectors through the Puffin format.
The actual measured performance and storage improvements depend on workload and environment and might differ from the preceding example.
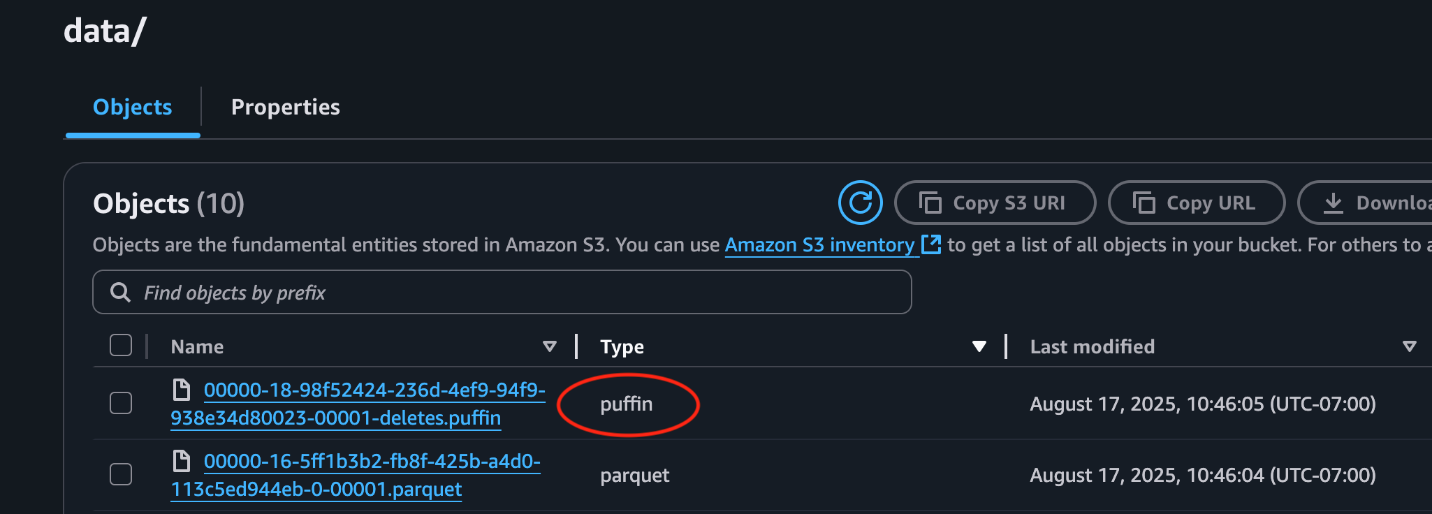
This following screenshot from the S3 bucket demonstrates a Puffin delete file stored alongside data files.
Clean up
After you finish your tests, it’s important to clean up your environment to avoid unnecessary costs:
- Drop the test tables you created to remove associated data from your S3 bucket and prevent ongoing storage charges.
- Delete any temporary data left in the S3 bucket used for Iceberg data.
- Delete the EMR cluster to stop billing for running compute resources.
Cleaning up resources promptly helps maintain cost-efficiency and resource hygiene in your AWS environment.
Considerations
Iceberg features are introduced through a phased process: first in the specification, then in the core library, and finally in engine implementations. Deletion vector support is currently available in the specification and core library, with Spark being the only supported engine. We validated this capability on Amazon EMR 7.10 with Spark 3.5.5.
Conclusion
Iceberg v3 introduces a significant advancement in managing row-level deletes for merge-on-read operations through binary deletion vectors stored in compact Puffin files. Our performance tests, conducted with Iceberg 1.9.2 on Amazon EMR 7.10.0 and EMR Spark 3.5.5, show clear improvements in both delete operation speed and read performance, along with a considerable reduction in delete file storage compared to Iceberg v2’s positional delete Parquet files. For more information about deletion vectors, refer to Iceberg v3 deletion vectors.