AWS Big Data Blog
Migrate terabytes of data quickly from Google Cloud to Amazon S3 with AWS Glue Connector for Google BigQuery
This blog post was last updated July, 2022 to update the new version of the connector and details on how to push down queries to Google BigQuery.
The cloud is often seen as advantageous for data lakes because of better security, faster time to deployment, better availability, more frequent feature and functionality updates, more elasticity, more geographic coverage, and costs linked to actual utilization. However, recent studies from Gartner and Harvard Business Review show multi-cloud and intercloud architectures are something leaders need to be prepared for as data management, governance, and integration become more complex. To make sure your data scientist has access to the right data to build their analytics processes, no matter where the data is stored, it’s imperative that moving terabytes of data from many different sources is simple, fast, and cost-effective.
The objective of this post is to describe how to use AWS Glue, a fully managed, serverless, extract, transform, and load (ETL) service that makes it easy to prepare and load your data for analytics,and the latest version of Google BigQuery Connector for AWS Glue to build an optimized ETL process to migrate a large and complex dataset from Google BigQuery storage into Amazon Simple Storage Service (Amazon S3), in Parquet format, in under 30 minutes.
The dataset used in the post is stored in the BigQuery table 1000_genomes_phase_3_variants_20150220, which collects open data by the IGRS and the 1000 Genomes Project. We selected this sample dataset for its size—1.94 TB and about 85 million rows—and because it has a complex schema with repeated attributes (nested structures), as shown in the following screenshot.

Solution overview
Our solution builds on top of the steps described in the post Migrating data from Google BigQuery to Amazon S3 using AWS Glue custom connectors. The following diagram illustrates our simplified architecture.

We further simplify the solution by using the new feature in AWS Glue Studio to create or update a table in the Data Catalog from the job. This removes the need to run an AWS Glue crawler before querying the data with Amazon Athena.
Prerequisites
Before getting started, make sure you meet the following prerequisites:
- Have an account in Google Cloud, specifically a service account that has permissions to Google BigQuery
- Complete the first three steps in the post Migrating data from Google BigQuery to Amazon S3 using AWS Glue custom connectors to configure your Google account, create an AWS Identity and Access Management (IAM) role (note down the name), and activate the BigQuery connector
Create the ETL job in AWS Glue Studio
To create your ETL job, complete the following steps:
- On the AWS Glue console, open AWS Glue Studio.
- In the navigation pane, choose Jobs.
- Choose Create job.
- Select Source and target added to the graph.
- For Source, choose Google BigQuery Connector 0.24.2 for AWS Glue 3.0 (choose Google BigQuery Connector 0.24.2 for AWS Glue 1.0 and 2.0 if you plan to use AWS Glue 1.0 or AWS Glue 2.0).
- For Target, choose S3.
- Choose Create.

- In the job editor, on the Visual tab, choose the node ApplyMapping.
- Choose Remove.

- Choose your data source node (Google BigQuery Connector for AWS Glue 3.0).
- On the Data source properties – Connector tab, for Connection, choose BigQuery.
- Under Connection options, choose Add new option.

You add two key-value pairs.
- For the first key pair, for Key, enter
parentProject, and for Value, enter your Google project name. - For the second key pair, for Key, enter table, and for Value, enter
bigquery-public-data.human_genome_variants.1000_genomes_phase_3_optimized_schema_variants_20150220.

- Choose your data target node (your S3 bucket).
- On the Data target properties – S3 tab, for Format¸ choose Glue Parquet.
This is a custom Parquet writer type that is optimized for DynamicFrames; a precomputed schema isn’t required before writing. The file in Amazon S3 is a standard Apache Parquet file.
- For S3 Target Location, enter the S3 path of your bucket.
- Select Create a table in the Data Catalog and on subsequent runs update the schema and add new partitions.
- For Database, choose the database you want your table in.
- For Table name, enter
1000_genomes_phase_3.
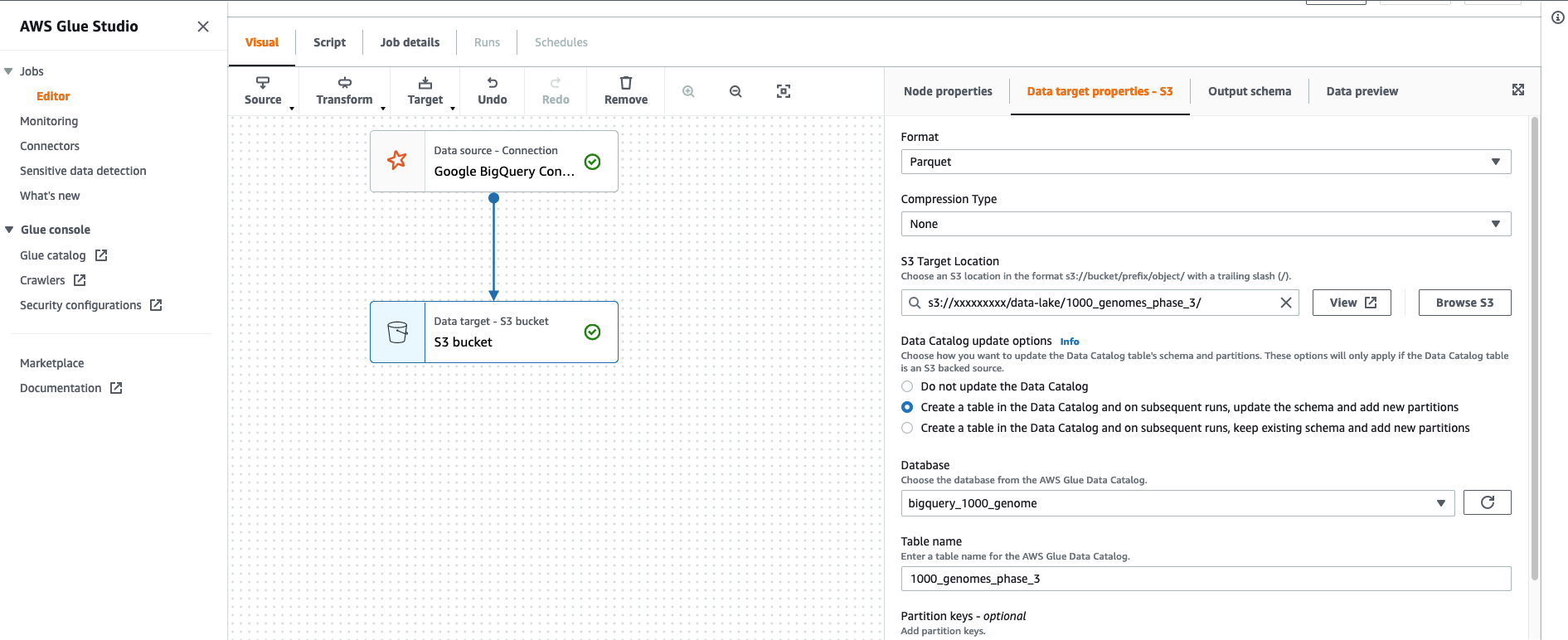
- Choose Save.
- On the Job details tab, for Name, enter
BigQuery_S3. - For IAM Role, choose the role you created as part of the prerequisite steps.
- For Type, choose Spark.
- For Glue version, choose Glue 3.0 – Supports Spark 3.1, Scala 2, Python3.
- To achieve maximum throughput migrating the data, enter
126for Number of workers.
By default, the connector creates one partition per 400MB in the table being read (before filtering). As of this writing, the Google Cloud BigQuery Storage API has a maximum of 1,000 parallel read streams. With the G.1X worker type chosen as default, each worker maps to 4 vCPU, 16 GB of memory, 64 GB disk, and provides one Spark executor. For more information, see Defining Job Properties. In Glue 2.0 with one node for the Spark driver and eight tasks per executor, the 126 workers enable us to run 1,000 tasks simultaneously. In Glue 3.0 each executor is configured to run 4 tasks, so we will run only 500 tasks simultaneously.
The following screenshot shows that the total run time was 13 minutes to migrate and convert 1.9 TB of data. We conducted the test with the AWS Glue job in the us-east-1 and eu-central-1 Regions, with the GCP data location set as US.

If you choose the job run id link, you can review the detailed metrics for the job. Looking at the Active Executors vs, the Maximum Needed Executors metric we notice that we could further improve the performance adding additional workers.

This is expected, in fact to reach the maximum BigQuery parallelism of 1000 we would need to double the number of tasks. If we run the job with 251 worker nodes it completes in just 8 minutes!

Validate the migration
To confirm the migration was successful, complete the following steps:
- On the AWS Glue console, under Databases, choose Tables.
- Search for the table
genomes_1000_phase_3.

- Choose the table and review the schema.
The repeated fields in BigQuery have been converted into an array in Parquet.
- Choose the S3 location link to open the Amazon S3 console.

The S3 location contains thousands of Parquet files, in average about 70 MB in size.
- To check the full size of the dataset (compared with the BigQuery size of 1.9 TB), choose the parent prefix
data_lake/in the S3 path.

- Select the object
genomes_1000_phase_3. - On the Actions menu, choose Calculate total size.

The total size in Amazon S3 is only 142.2 GB—7.48% of the total volume of data in BigQuery storage. This is because in Amazon S3, we store the data in Apache Parquet with Snappy encoding, an open columnar file format that allows us to achieve high compression ratios. In contrast, Google BigQuery data size is calculated based on the size of each column’s data type, even if the data is internally compressed.

To validate that all the data has been successfully migrated, we run two queries in Athena.
- On the Athena console, run the following query:
The results show that the amount of records match what we have in BigQuery: 84,801,880.

- Run a second query:
In this query, Athena only needs to scan 212.93 KB of data instead of 274.1 MB (as with BigQuery). Consider the implications of a full table scan from a cost perspective (142.2 GB of data scan vs, 1,900 GB).

Scale out
If your tables are partitioned or even larger and you want to split the migration into multiple jobs, you can pass two additional parameters to your connection options:
- filter – Passes the condition to select the rows to convert. If the table is partitioned, the selection is pushed down and only the rows in the specified partition are transferred to AWS Glue. In all other cases, all data is scanned and the filter is applied in AWS Glue Spark processing, but it still helps limit the amount of memory used in total.
- maxParallelism – Defines the number of read streams used to query the BigQuery storage. This can help you avoid exhausting your BigQuery storage quota of 5,000 read rows per project per user per minute if you want to submit multiple jobs in parallel.

For example, in the following screenshot, our workflow ran 10 jobs in parallel on our source table, with maxParallelism set to 500 and filter configured to a set of values for reference_name that selects approximately 1/10th of the rows in the table (8 million rows). The workflow completed in 19 minutes.

In the latest version of the connector, you can replace the filter option with the SQL query you want to run on BigQuery. Notice that the execution should be faster as only the result is transmitted over the wire.
In order to use this feature the following configurations MUST be set:
viewsEnabledmust be set to true.materializationDatasetmust be set to a dataset where the GCP user has table creation permission.materializationProjectis optional.
In the AWS Glue Studio job, the connection options to set are:
- query – Pushes down the SQL query for execution, selecting a subset of columns or adding a filter clause reduce the data scanned by Google BigQuery, optimizing performance and costs.
- viewsEnabled – Enables the connector to read from views and not only tables. Set it to true.
- materializationDataset – The dataset where the materialized view is going to be created. You can provide the value of an existing dataset or a new one.
Pricing considerations
You might have egress charges for migrating data out of Google BigQuery into Amazon S3. Review and calculate the cost for moving your data. As of this writing, AWS Glue 3.0 charges $0.44 per DPU-hour, billed per second, with a 1-minute minimum for Spark ETL jobs. The jobs in our test were configured with 126 DPUs and ran in 13 minutes; the total runtime was 25.39 DPU-hours for a cost of approximately $11. For more information, see AWS Glue pricing. With Auto Scaling enabled, AWS Glue automatically adds and removes workers from the cluster depending on the parallelism at each stage or microbatch of the job run. You no longer need to worry about over-provisioning resources for jobs, spend time optimizing the number of workers, or pay for idle workers. As you can see from the following screenshot of the Job runs table in the Glue Studio Monitoring tab. It shows how the job run with 251 nodes completed in 8 minutes, but consumed only 28.79 DPU hours instead of the expected 33.46.

To avoid incurring future charges, delete the data in the S3 buckets. If you’re not running an ETL job or crawler, you’re not charged. Alternatively, you can delete the AWS Glue ETL job and Data Catalog tables.
Conclusion
In this post, we learned how to easily customize an AWS Glue ETL job that connects to a BigQuery table and migrates a large amount of data (1.9 TB) into Amazon S3, quickly (about 8 minutes). We then queried the data with Athena to demonstrate the savings in storage costs (90% + compression factor).
With AWS Glue, you can significantly reduce the cost, complexity, and time spent creating ETL jobs. AWS Glue is serverless, so there is no infrastructure to set up or manage. You pay only for the resources consumed while your jobs are running.
For more information about AWS Glue ETL jobs, see Simplify data pipelines with AWS Glue automatic code generation and workflows and Making ETL easier with AWS Glue Studio.
About the Author
 Fabrizio Napolitano is a Senior Specialist SA for DB and Analytics. He has worked in the analytics space for the last 20 years, and has recently and quite by surprise become a Hockey Dad after moving to Canada.
Fabrizio Napolitano is a Senior Specialist SA for DB and Analytics. He has worked in the analytics space for the last 20 years, and has recently and quite by surprise become a Hockey Dad after moving to Canada.

