AWS Big Data Blog
Create data science environments on AWS for health analysis using OHDSI
Applying technology to healthcare data has the potential to produce many exciting and important outcomes. The analysis produced from healthcare data can empower clinicians to improve the health of individuals and populations by enabling them to make better decisions that enhance the care they provide.
The Observational Health Data Sciences and Informatics (OHDSI) program and community are working toward this goal by producing data standards and open-source solutions to store and analyze observational health data. Using the OHDSI tools, you can visualize the health of your entire population. You can build cohorts of patients, analyze incidence rates for various conditions, and estimate the effect of treatments on patients with certain conditions. You can also model health outcome predictions using machine learning algorithms.
One of the challenges often faced when working with big data tools is the expense of the infrastructure required to run them. Another challenge is the learning curve to implement and begin using these tools. AWS makes it possible to address many of the classic IT challenges by making enterprise-class infrastructure and technology available in an affordable, elastic, and automated way.
The OHDSIonAWS project is automation that combines more than a dozen of the OHDSI projects (Atlas, PatientLevelPrediction, Achilles, the OMOP Common Data Model, and more) with AWS technologies. Using it, you can quickly and inexpensively implement a health data science and informatics environment.
This post outlines the features of the OHDSIonAWS architecture and helps you to get started using it in your AWS account.
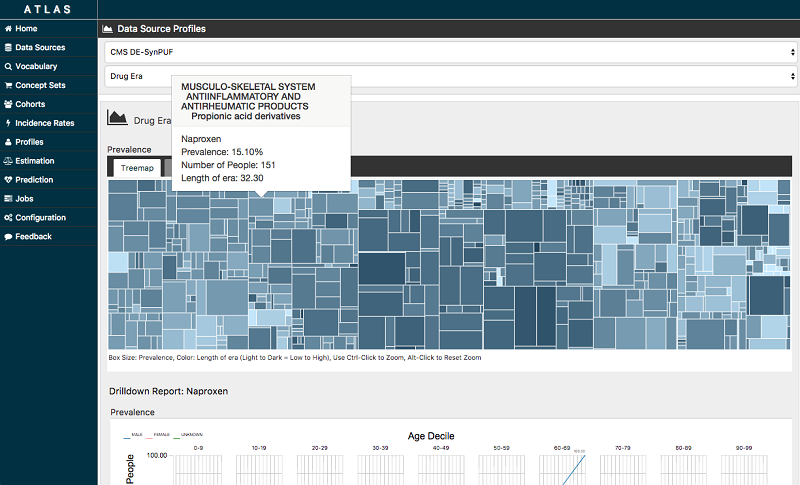
The following screenshot is just one example of the population health analysis that is possible with the OHDSI tools. This Atlas visualization shows the prevalence of various drugs within the given population of people. This information helps researchers and clinicians discover trends and make informed decisions about patient health.
OHDSIonAWS application architecture
Before deploying an application on AWS that transmits, processes, or stores protected health information (PHI) or personally identifiable information (PII), address your organization’s compliance concerns. Work with your internal compliance and legal team to ensure compliance with the laws and regulations that govern your organization. To understand how you can use AWS services as a part of your overall compliance program, see the AWS HIPAA Compliance whitepaper.
Enterprise organizations can use OHDSIonAWS to create an observational health data analytics environment using the OMOP Common Data Model and the OHDSI data science toolset. For more information about OHDSI personal learning or training environments, see the OHDSI-in-a-Box project.
This post gives an overview of the OHDSIonAWS project, which deploys a complete enterprise OHDSI environment on AWS, including a data warehouse with synthetic sample data. It has the following features:
- Deploys an enterprise OHDSI environment automatically, configured with the latest versions of more than a dozen OHDSI tools in just a few hours.
- Uses deployment parameters to configure environments from small to petabyte-scale, geographically redundant implementations.
- Includes several options for public, synthetic, sample datasets. Can also load your data private datasets automatically.
- Deploys with internet access, or can be accessible only from within your organization’s private network.
- Provides role-based access control.
- Deploys in an isolated, three-tier VPC with high availability.
- Uses data-at-rest and in-flight encryption.
- Uses managed services from AWS. OS, middleware, and database patching and maintenance are largely automatic.
- Creates automated backups for operational and disaster recovery.
- Produces a low monthly cost compared to comparable on-premises architectures.
The following diagram illustrates how the OHDSI tools map to AWS services.
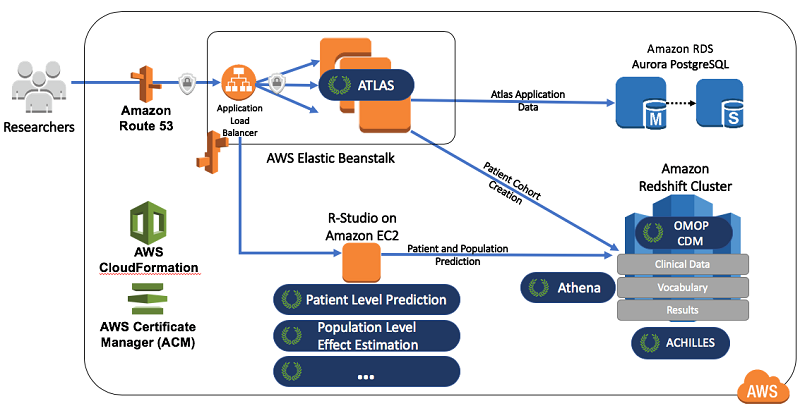
Researchers interact with the web application Atlas to perform analysis. In this example, AWS Elastic Beanstalk deploys and manages Atlas. Elastic Beanstalk is an easy-to-use service for deploying and scaling web applications.
After receiving the compiled Atlas code, Elastic Beanstalk automatically handles the deployment of the web application infrastructure. It covers everything from capacity provisioning, load balancing, automatic scaling, and high availability, to application health monitoring. Using a feature of Elastic Beanstalk called ebextensions, you can customize the Atlas servers to use an encrypted storage volume for the middleware application logs.
Atlas stores the state of the various patient cohorts that a dedicated database analyzes, separate from your observational health data. Amazon Aurora provides this database with PostgreSQL compatibility.
Aurora is a relational database built for the cloud that combines the performance and availability of high-end commercial databases with the simplicity and cost-effectiveness of open-source databases. It provides cost-efficient and resizable capacity while automating time-consuming administration tasks such as hardware provisioning, database setup, patching, and backups. It is configured for high availability and uses encryption at rest for the database and backups, and encryption in flight for the JDBC connections.
Through the parameters that you provide OHDSIonAWS, you can specify a single-instance, highly available, or serverless implementation of Aurora PostgreSQL. You can accomplish this through the following AWS CloudFormation code:
The OHDSI Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) stores all your observational health data. This model also stores useful vocabulary tables that help to translate values from various data sources (like electronic health record systems and claims data).
The OMOP CDM schema deploys onto Amazon Redshift. Amazon Redshift is a fast, fully managed data warehouse that allows you to run complex analytic queries against petabytes of structured data. It uses sophisticated query optimization, columnar storage on high-performance local disks, and massively parallel query execution. You can also resize an Amazon Redshift cluster as your requirements change.
RStudio Server comes with the OHDSI Patient Level Prediction R library pre-installed and ready to use, as well as more than a dozen other OHDSI R libraries. You can use these R libraries to analyze the patient-level data and cohorts managed by Atlas.
Use Amazon EC2 to implement RStudio Server Open Source Edition. EC2 provides a web-based IDE for R development. The user accounts that provide access to RStudio are created locally on the EC2 server. The Professional Edition of RStudio, available in the AWS Marketplace, provides the ability to integrate with external identity repositories like Active Directory or LDAP, which your organization may find useful.
On the RStudio instance, you can mount an encrypted Amazon EBS volume as the home directory for all RStudio users’ files. This encryption respects the HIPAA requirement for encryption at rest of PHI. You can implement the encryption with the following CloudFormation code:
The following detailed technical diagram shows the configuration of the deployed architecture. You can deploy the OHDSIonAWS architecture so that it’s accessible from the internet, or only through your organization’s internal, private network. For private network deployments, you can connect your private network in AWS to your on-premises, internal network using a VPN connection or an AWS Direct Connect connection.
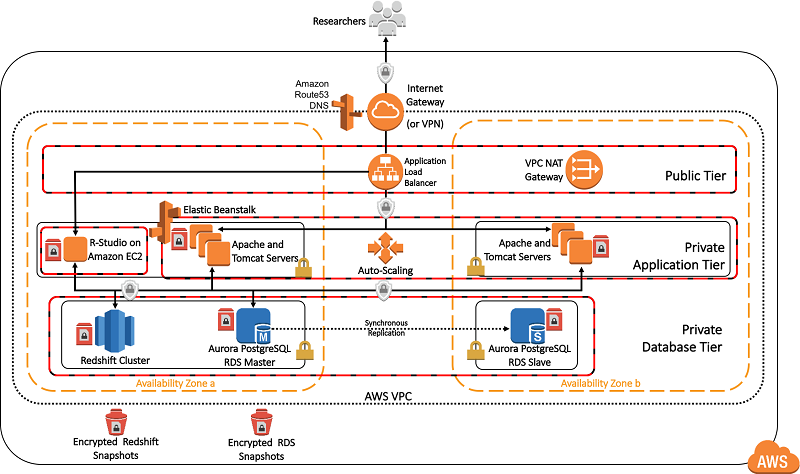
Deploying OHDSI on AWS
Select the appropriate Launch Stack button to begin deploying your OHDSIonAWS environment. For more geographic Region choices, see the OHDSIonAWS GitHub repository.
|
AWS Region Name |
Launch internet Accessible Environment |
Launch Private Network Only Environment |
| US East (N. Virginia) |  |
 |
| US West (N. California) |  |
 |
| EU (Ireland) |  |
 |
| EU (Frankfurt) |  |
 |
For detailed instructions on the initial deployment and ongoing operation of the OHDSIonAWS architecture, see the OHDSIonAWS GitHub repository. Highlights of some of the key parameter configurations of this architecture are as follows:
- DNS and SSL – Automatically deploys an Amazon Route 53 DNS name and SSL certificate for your OHDSIonAWS environment. This function enables encrypted, HTTPS communication to Atlas and RStudio, helping you to respect the requirement of HIPAA that you must encrypt PHI in transit.
- Database Tier – Configures the OHDSI Atlas database as either standalone, highly available, or serverless using the Aurora PostgreSQL database technology. These parameters also let you specify the size and type of Amazon Redshift cluster, which allows you to scale up the capacity and performance of the data warehouse that holds your OMOP Common Data Model data.
- OMOP Sources – Specifies the OMOP-formatted data sources that automatically load into the OHDSIonAWS environment. These sources are also automatically characterized by OHDSI Achilles after they are loaded. The OHDSIonAWS architecture allows you to choose from six public, synthetic data sources, or you can load private OMOP data sources. The following is a table of the synthetic data sources available.
|
Sample Data Source |
Size |
Schema Name |
| Synthea | 1,000 persons | synthea1k |
| Synthea | 100,000 persons | synthea100k |
| Synthea | >2,000,000 persons | synthea23m |
| CMS DeSynPUF | 1,000 persons | CMSDESynPUF1k |
| CMS DeSynPUF | 100,000 persons | CMSDESynPUF100k |
| CMS DeSynPUF | >2,000,000 persons | CMSDESynPUF23m |
- RStudio – Specify the size and type of EC2 instance that is to host RStudio Server as well as the OHDSI R libraries like OHDSI PatientLevelPrediction. This instance can include GPUs to speed the training of machine learning models if you choose. You also provide a comma-delimited list of user names and passwords that are used to access the RStudio environment. You can change passwords and add or deploy users by following the instruction on the OHDSIonAWS GitHub repository.
- Atlas – Specify the size and type of EC2 instances that host the OHDSI Atlas web interface. These instances are configured in an AWS Auto Scaling group that automatically adds and removes instances based on load. You can also define the minimum and maximum number of instances in this Auto Scaling group. The instances are automatically spread across multiple Availability Zones. As long as your minimum is at least two, you have a high-available configuration. Optionally, you can also choose to apply the user names and passwords you supplied for RStudio to enforce role-based access to Atlas.
- OHDSI Project Versions – Specify the versions of more than a dozen OHDSI projects that deploy into your OHDSIonAWS environment. These OHDSI projects are dynamically built during deployment from their release branches in GitHub, allowing you to create repeatable and reproducible OHDSI environments. As new versions of OHDSI projects are released, you can easily specify new versions to create test environments before updating your production environment.
- VPC Networking – Specify the VPC configuration into which to deploy your OHDSIonAWS environment. For the internet-accessible version, you can optionally specify the network address ranges to use. A three-tiered VPC with security groups, secure routes, and internet access is then configured and deployed automatically. For the Private Network Only version, specify a pre-existing VPC and subnets into which to deploy your OHDSIonAWS environment. This feature enables you to use a VPC that is already configured with connectivity back to your internal, private network.
After you provide these parameters, your OHDSIonAWS environment automatically deploys over a few hours, depending on the size of your data sources and scale of your Amazon Redshift configuration.
Cost of deploying this environment
It used to be common to see healthcare data analytics environments deployed in an on-premises data center with expensive data warehouse appliances and virtualized environments. The cloud era has democratized the availability of the infrastructure required to do this type of data analysis, so that now it is within reach of even small organizations.
This environment can expand to analyze petabyte-scale health data, and you only pay for what you need. For an estimated breakdown of the monthly cost components for this environment as deployed, see OHDSI on AWS Architecture on the AWS Solution Calculator.
You do not have to run this environment all of the time. If you only perform analyses periodically, you can terminate the environment when you are finished and restore it from the database backups when you want to continue working. This feature further reduces the cost of operation.
Summary
Now that you have a fully functional OHDSI environment with sample data, you can use this to explore and learn the toolset and its capabilities. After learning with the sample data, you can gain insights by analyzing your own organization’s health data.
The OHDSI toolset and standards enable broad-scale, reproducible research of phenotypic, observational health data. The free and open-source nature of OHDSI means that these capabilities are open to organizations of any size. The agility, scalability, and low cost of AWS infrastructure enables the extensive and consistent use of OHDSI without significant capital investment or lengthy IT efforts.
Additional Reading
If you found this post useful, be sure to check out Build a Healthcare Data Warehouse Using Amazon EMR, Amazon Redshift, AWS Lambda, and OMOP for info on how to automate data ETL to the OMOP CDM.

About the Author
 James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.
James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.