AWS Big Data Blog
Build a Healthcare Data Warehouse Using Amazon EMR, Amazon Redshift, AWS Lambda, and OMOP
In the healthcare field, data comes in all shapes and sizes. Despite efforts to standardize terminology, some concepts (e.g., blood glucose) are still often depicted in different ways. This post demonstrates how to convert an openly available dataset called MIMIC-III, which consists of de-identified medical data for about 40,000 patients, into an open source data model known as the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM). It describes the architecture and steps for analyzing data across various disconnected sources of health datasets so you can start applying Big Data methods to health research.
Before designing and deploying a healthcare application on AWS, make sure that you read through the AWS HIPAA Compliance whitepaper. This covers the information necessary for processing and storing patient health information (PHI).
Note: If you arrived at this page looking for more info on the movie Mimic 3: Sentinel, you might not enjoy this post.
OMOP overview
The OMOP CDM helps standardize healthcare data and makes it easier to analyze outcomes at a large scale. The CDM is gaining a lot of traction in the health research community, which is deeply involved in developing and adopting a common data model. Community resources are available for converting datasets, and there are software tools to help unlock your data after it’s in the OMOP format. The great advantage of converting data sources into a standard data model like OMOP is that it allows for streamlined, comprehensive analytics and helps remove the variability associated with analyzing health records from different sources.
OMOP ETL with Apache Spark
Observational Health Data Sciences and Informatics (OHDSI) provides the OMOP CDM in a variety of formats, including Apache Impala, Oracle, PostgreSQL, and SQL Server. (See the OHDSI Common Data Model repo in GitHub.) In this scenario, the data is moved to AWS to take advantage of the unbounded scale of Amazon EMR and serverless technologies, and the variety of AWS services that can help make sense of the data in a cost-effective way—including Amazon Machine Learning, Amazon QuickSight, and Amazon Redshift.
This example demonstrates an architecture that can be used to run SQL-based extract, transform, load (ETL) jobs to map any data source to the OMOP CDM. It uses MIMIC ETL code provided by Md. Shamsuzzoha Bayzid. The code was modified to run in Amazon Redshift.
Getting access to the MIMIC-III data
Before you can retrieve the MIMIC-III data, you must request access on the PhysioNet website, which is hosted on Amazon S3 as part of the Amazon Web Services (AWS) Public Dataset Program. However, you don’t need access to the MIMIC-III data to follow along with this post.
Solution architecture and loading process
The following diagram shows the architecture that is used to convert the MIMIC-III dataset to the OMOP CDM.
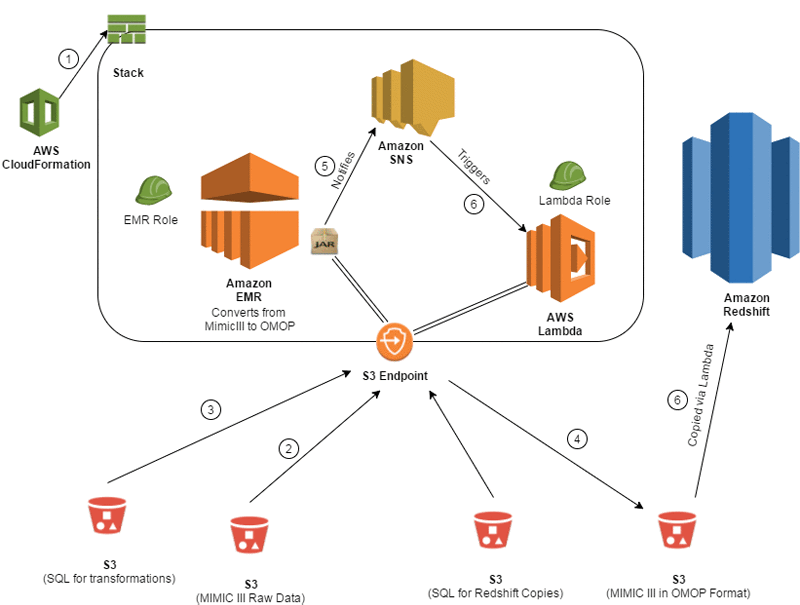
The data conversion process includes the following steps:
- The entire infrastructure is spun up using an AWS CloudFormation template. This includes the Amazon EMR cluster, Amazon SNS topics/subscriptions, an AWS Lambda function and trigger, and AWS Identity and Access Management (IAM) roles.
- The MIMIC-III data is read in via an Apache Spark program that is running on Amazon EMR. The files are registered as tables in Spark so that they can be queried by Spark SQL.
- The transformation queries are located in a separate Amazon S3 location, which is read in by Spark and executed on the newly registered tables to convert the data into OMOP form.
- The data is then written to a staging S3 location, where it is ready to be copied into Amazon Redshift.
- As each file is loaded in OMOP form into S3, the Spark program sends a message to an SNS topic that signifies that the load completed successfully.
- After that message is pushed, it triggers a Lambda function that consumes the message and executes a COPY command from S3 into Amazon Redshift for the appropriate table.
This architecture provides a scalable way to use various healthcare sources and convert them to OMOP format, where the only changes needed are in the SQL transformation files. The transformation logic is stored in an S3 bucket and is completely de-coupled from the Apache Spark program that runs on EMR and converts the data into OMOP form. This makes the transformation code portable and allows the Spark jar to be reused if other data sources are added—for example, electronic health records (EHR), billing systems, and other research datasets.
Note: For larger files, you might experience the five-minute timeout limitation in Lambda. In that scenario you can use AWS Step Functions to split the file and load it one piece at a time.
Scaling the solution
The transformation code runs in a Spark container that can scale out based on how you define your EMR cluster. There are no single points of failure. As your data grows, your infrastructure can grow without requiring any changes to the underlying architecture.
If you add more data sources, such as EHRs and other research data, the high-level view of the ETL would look like the following:
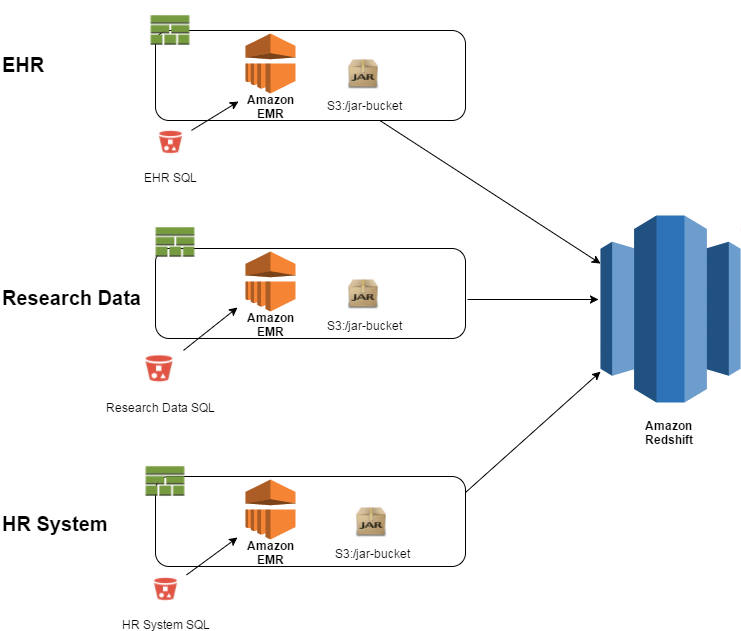
In this case, the loads of the different systems are completely independent. If the EHR load is four times the size that you expected and uses all the resources, it has no impact on the Research Data or HR System loads because they are in separate containers.
You can scale your EMR cluster based on the size of the data that you anticipate. For example, you can have a 50-node cluster in your container for loading EHR data and a 2-node cluster for loading the HR System. This design helps you scale the resources based on what you consume, as opposed to expensive infrastructure sitting idle.
The only code that is unique to each execution is any diffs between the CloudFormation templates (e.g., cluster size and SQL file locations) and the transformation SQL that resides in S3 buckets. The Spark jar that is executed as an EMR step is reused across all three executions.
Upgrading versions
In this architecture, upgrading the versions of Amazon EMR, Apache Hadoop, or Spark requires a one-time change to one line of code in the CloudFormation template:
Note that this example uses a slightly lower version of EMR so that it can use Spark 2.0.0 instead of Spark 2.1.0, which does not support nulls in CSV files.
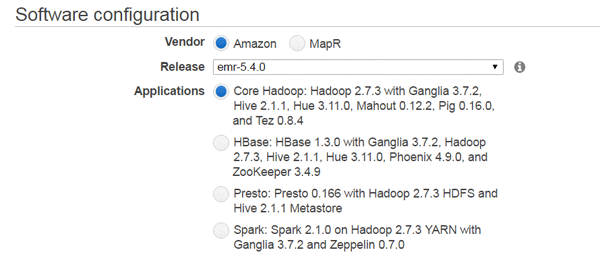
You can also select the version in the Release list in the General Configuration section of the EMR console:
The data sources all have different CloudFormation templates, so you can upgrade one data source at a time or upgrade them all together. As long as the reusable Spark jar is compatible with the new version, none of the transformation code has to change.
Executing queries on the data
After all the data is loaded, it’s easy to tear down the CloudFormation stack so you don’t pay for resources that aren’t being used:
This includes the EMR cluster, Lambda function, SNS topics and subscriptions, and temporary IAM roles that were created to push the data to Amazon Redshift. The S3 buckets that contain the raw MIMIC-III data and the data in OMOP form remain because they existed outside the CloudFormation stack.
You can now connect to the Amazon Redshift cluster and start executing queries on the ten OMOP tables that were created, as shown in the following example:
OMOP analytics tools
For information about open source analytics tools that are built on top of the OMOP model, visit the OHDSI Software page.
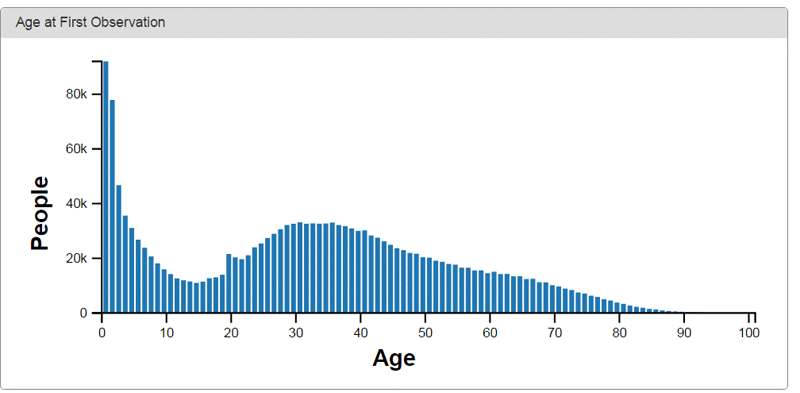
The following are examples of data visualizations provided by Achilles, an open source visualization tool for OMOP.
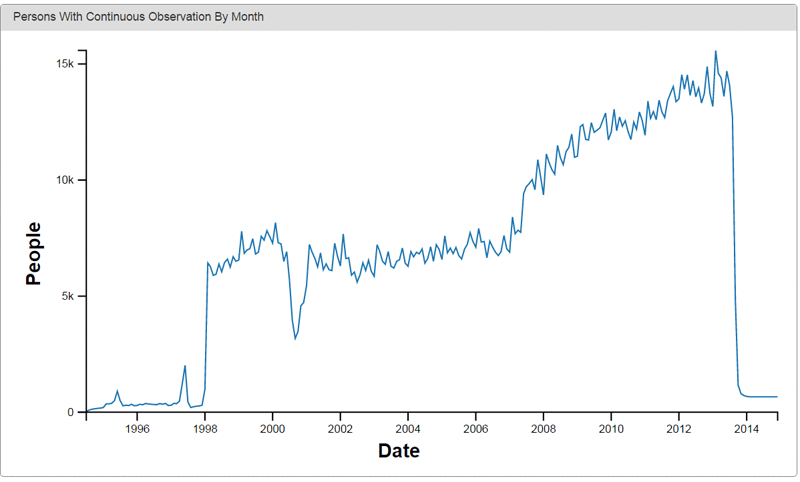
Conclusion
This post demonstrated how to convert MIMIC-III data into OMOP form using data tools that are built for scale and flexibility. It compared the architecture against a traditional data warehouse and showed how this design scales by mixing a scale-out technology with EMR and a serverless technology with Lambda. It also showed how you can lower your costs by using CloudFormation to create your data pipeline infrastructure. And by tearing down the stack after the data is loaded, you don’t pay for idle servers.
You can find all the code in the AWS Labs GitHub repo with detailed, step-by-step instructions on how to load the data from MIMIC-III to OMOP using this design.
If you have any questions or suggestions, please add them below.
About the Author
 Ryan Hood is a Data Engineer for AWS. He works on big data projects leveraging the newest AWS offerings. In his spare time, he enjoys watching the Cubs win the World Series and attempting to Sous-vide anything he can find in his refrigerator.
Ryan Hood is a Data Engineer for AWS. He works on big data projects leveraging the newest AWS offerings. In his spare time, he enjoys watching the Cubs win the World Series and attempting to Sous-vide anything he can find in his refrigerator.
Related
Create a Healthcare Data Hub with AWS and Mirth Connect
