How was this content?
CelerisTx: Drug Discovery for Incurable Diseases with ML on AWS
Harnessing computational drug discovery on proximity-inducing compounds (PICs)
Humanity forged ahead with countless therapeutic solutions to treat disease. Recent technologies such as knockout (CRISPR – works at the DNA level) and knockdown (RNAi – works at the RNA level) have significant limitations. In contrast, Proximity-Inducing Compounds (PICs) combine the unique advantages of small molecules. Such beneficial features include oral bioavailability and ease of manufacture – along with those of silencing technologies such as CRISPR and RNAi. In contrast to RNAi and CRISPR, PICs affect proteins, thus providing a breakthrough therapeutic modality to target proteins, specifically associated with diseases. Alternatives that target proteins include biologics and inhibitors, and these alternatives have not expanded the range of pathogenic proteins that can be targeted. Over 80% of pathogenic proteins are associated with a disease not yet amenable to pharmaceutical interventions.
Targeted protein degradation (TPD) is the most popular proximity-inducing drug modality, first demonstrated in the early 2000s. It also received a Nobel Prize in Chemistry for discovering ubiquitin-mediated protein degradation in 2004. The TPD concept involves the selective degradation of proteins by hijacking internal cellular machinery within the human body. Compared to occupancy-driven methods such as inhibitors, TPD mechanisms have the advantage of eliminating scaffolding functions and thus addressing the root cause of the problem rather than just treating symptoms. In general, tools to eliminate misfolded and denatured proteins within a cell are needed to maintain biological homeostasis.
While various machineries can be actively utilized via PICs, such as lysosomes and autophagy, Celeris Therapeutics focuses on the ubiquitin-proteasome system. Based on this system, unwanted proteins are tagged with ubiquitin, a small signaling protein. As a result, this process initiates the degradation of pathogenic proteins.
However, the assessment for TPD in an experimental laboratory setup is slow and expensive. Celeris Therapeutics, therefore, implemented a computational workflow to predict protein degradation effectively and thus shorten and streamline the drug development timeline.
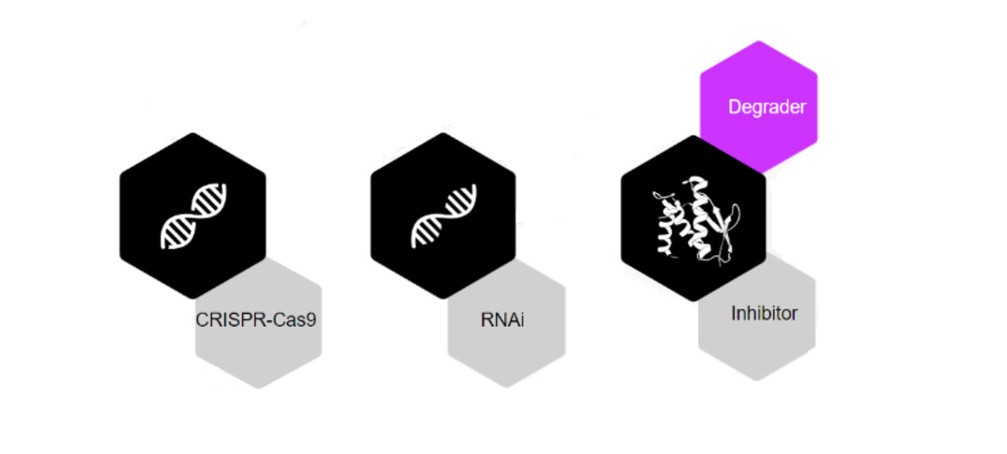
Proximity-inducing compounds
Celeris Therapeutics (CelerisTx) is pioneering the adoption of AI on PICs, focusing on Targeted Protein Degradation. In terms of data, in protein degradation, that means identifying multiple molecules that interact with each other in a particular way. Imagine LEGO® bricks. Instead of fitting them together according to only a single dimension i.e., shape, these elements have multiple dimensions or requirements to satisfy. Some are chemical, biological, physical-driven, etc., meaning, that the interactions are subject to specific laws. These laws need to be quantified and embedded in data. Concrete examples of such information include 3D structures of molecules represented as graphs, hydrophobicity, or electrostatic potential. Altogether, this amounts to roughly 20 different features that experts in TPD curate. As a result, an enormous amount of data is being generated that needs to be analyzed using machine learning algorithms and, specifically, geometric deep learning pipelines. A concrete ML application is required to determine the degree of interaction between molecules involved in a so-called ternary complex. In other words, considering two proteins, we expect a scalar value that determines the degree of interaction. Beyond being accurate and generalizable, we have to make rapid predictions in a performant system. As we simulate different protein-protein interactions, it is necessary to discard inaccurate predictions swiftly to parse through the space promptly. This has proven to be a challenge for an interaction dataset with more than 20,000 protein pairs.
How AWS was leveraged for various PIC discovery projects
To save developer time and gain quicker insight into the particular problems surrounding machine learning development, we turned to Amazon SageMaker. Its capabilities allowed us to avoid implementing some machine learning infrastructure solutions ourselves, such as bias detection or hyperparameter tuning. Bias detection was relevant during data preparation; hence, we leveraged SageMaker Clarify. This was instrumental in ensuring the quality of our data, before we even started modeling. Afterward, once we had begun modeling, it was important to have direct integrations of geometric deep learning libraries, such as SageMaker Deep Graph Library (DGL). Since DGL is an open-source python package for deep learning on graphs, we used it to gain a quick setup of the infrastructure needed for geometric deep learning, which is essential.
Once the initial models were set, to find the best hyperparameters quickly, we used SageMaker Automatic Model Tuning. We estimate that we thereby avoided months of development time in coding up the hyperparameter optimization frameworks. We leveraged SageMaker Experiments to track and organize all changes to the experiments. This solution was especially important. Consider the aforementioned problem of determining the strength of the interaction between proteins involved in the ternary complex, the 15+ geometric deep learning architectures that we have experimented with from attention layers, the pure convolution operators on graphs, etc. The trackability of the different models and their respective parameters was important, to determine what would eventually work. Once we had created the final version of the model, we needed to debug them and further optimize the pipeline. For this, SageMaker Debugger proved helpful.
Along with software solutions from SageMaker, we needed a great deal of computing power for our vast data and deep learning pipelines. Hence, we built an optimized Spot Fleet solution for the GPUs used in training our machine learning workflows.
Optimizing the cost of our ML pipeline
Spot Instances are a special kind of Amazon EC2 instance that give 90% discount compared to On-Demand prices. Sometimes, due to high demand for computing resources, computing resources can be interrupted, causing a user to lose a whole compute session. It is crucial to save the computation’s intermediate results, if one trains a deep learning model for a long time, or if some necessarily long-running computation was executed on the Spot Instance. Aside from saving the computation’s intermediate results, the user should also be able to automatically continue the computation on another Spot Instance from the same intermediate step where the disconnect happened.
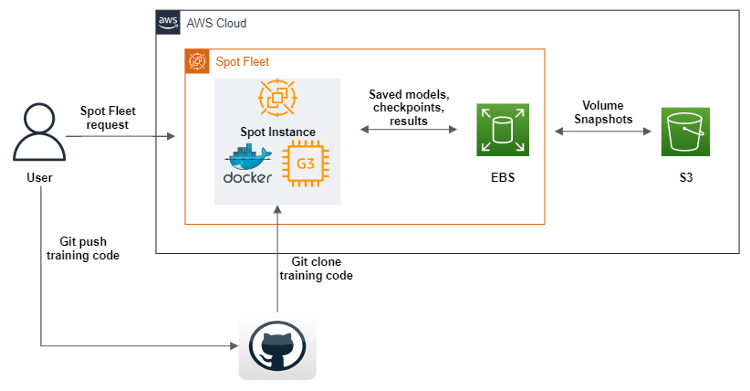
The AWS SDK for Python (boto3) was used to create a script that will, upon execution, send a Spot Fleet request to create a GPU Spot Instance with Amazon Machine Images (AMI), containing all of the software and dependencies to execute our machine learning code. Further, we define user data scripts that download a docker image from Amazon Elastic Container Registery (Amazon ECR), and then Git clones the code from our repository. Finally, the user data scripts executes the geometric deep learning pipeline code. This code has been modified such that after each epoch of machine learning training, it sends datasets, logs, models, and checkpoints to an Amazon EBS volume. To ensure that the objects are saved, we dump them all to Amazon S3. Aside from defining a Spot Fleet, we have also enabled replenishing any Spot Instances that disconnect in the process, which allows us to continue the training automatically from the last checkpoints found in the S3 buckets.
SageMaker offers something very similar, called Managed Spot Training. However, we did not leverage the SageMaker offering, because, when used as part of SageMaker, the EC2 instances are more expensive than standard ones. This makes sense, since SageMaker provides a lot of ML offerings, ease of use, less engineering time needed, etc. For us, architecting this solution was a strategic investment of time, because we knew we were going to use GPUs in different capacities for multiple years.
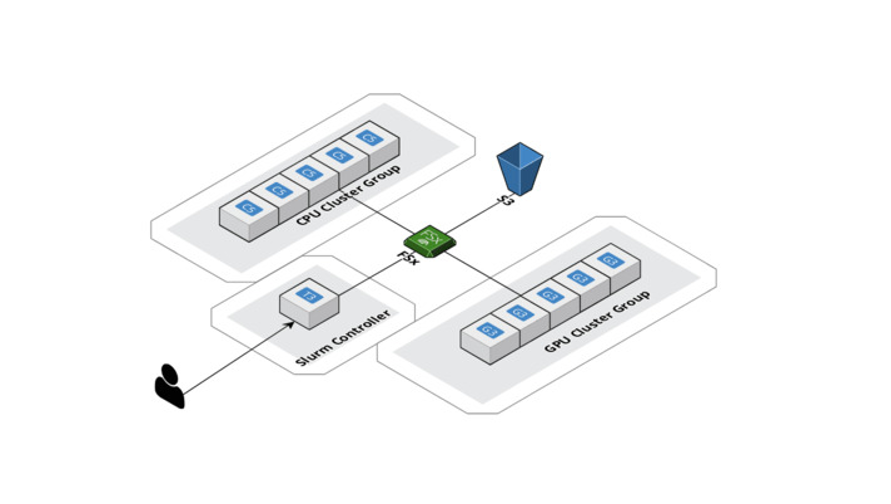
HPC was another significant use case that we needed to fulfill with AWS. The huge data space that needs to be parsed with geometric deep learning comes with a substantial computational burden. We scaled up our computations horizontally with AWS ParallelCluster and created a shared filed system, Amazon FSx for Lustre, so that all compute nodes could access and modify data from the same place. Also, AWS supports Slurm as a workload manager, which allows us to distribute the work across CPUs and GPUs at the same time. This eventually allows not only faster experimentation but also optimized model serving. Again, one should note that SageMaker offers a similar service for large datasets and models, called Distributed Training Libraries.
The future of proximity-inducing compounds
Preliminary results from our pipeline are available for Parkinson’s disease progressing to preclinical studies. Experimenting quickly and reliably is essential for computational drug discovery as we continue to improve our drug discovery pipeline with new architectures and approaches. Our R&D approach, in which we invest substantial resources in research, means that we face many ongoing risks we need to mitigate. AWS standard offerings are one way to ensure we are agile and move faster from research to lab and then to market.
Conclusion
Now is the time to increase productivity in drug discovery. The inverse of Moore’s Law in pharmacology, known as “Eroom’s Law,” indicates research and development costs required to develop a new drug will only continue to rise. An approach to address this is to harness and streamline the process of performing sustained machine learning experiments for drug discovery.

Christopher Trummer
Christopher Trummer is a Co-Founder of Celeris Therapeutics and serves as CEO. He has been an invited keynote speaker in AI for drug discovery conferences multiple times and is co-author of peer-reviewed publications in various journals.

Noah Weber
Noah Weber serves as Chief Technology Officer at Celeris Therapeutics. He is a Kaggle Grandmaster and adjunct lecturer at the Vienna University of Technology and the Vienna University of Applied Sciences.

Olajide Enigbokan
Olajide Enigbokan is a Startup Solutions Architect at Amazon Web Services. He loves working with startups (most especially builders) to discover the value of the AWS cloud.

AWS Editorial Team
The AWS Startups Content Marketing Team collaborates with startups of all sizes and across all sectors to deliver exceptional content that educates, entertains, and inspires.
How was this content?