AWS Startups Blog
Scaling High-Throughput Genomics Workflows on AWS
Guest post by Tomaz Berisa, Cofounder and CTO at Gencove
We have been working hard to scale low-pass sequencing at Gencove and ran into a few computational scalability constraints. This is an overview of how we got around them and the resulting architecture, complete with infrastructure templates and code.
Sequencing the first human genome in the early 2000s was an enormous success, but a costly undertaking valued at around ~$300 MM. The price of deeply sequencing a human genome has since been dropping dramatically (~$1000 today), enabling geneticists to sequence many more genomes and start characterizing the impact of genetics on characteristics like height, predisposition to disease, etc.
Although $1000 per deeply sequenced genome represents unprecedented progress, it is still expensive to sequence the tens or hundreds of thousands of genomes needed to learn about the genetics of many human traits and diseases.
At Gencove, we’ve developed low-pass sequencing technology, which relies on doing less sequencing and more compute to bring the price of a genome down to tens of dollars. We sequence a random subset (typically 40% or “0.4X”) of the genome and fill in the blanks by pattern-matching with large collections of genomic data using a method called imputation¹. This approach results in a powerful² sequencing product at the price point of legacy technology, i.e., genotyping arrays.
Gencove makes low-pass sequencing technology available as a platform, which packs all the complexities of sequencing genomes and generating genomic data ready for custom downstream analyses into a simple service behind an API:
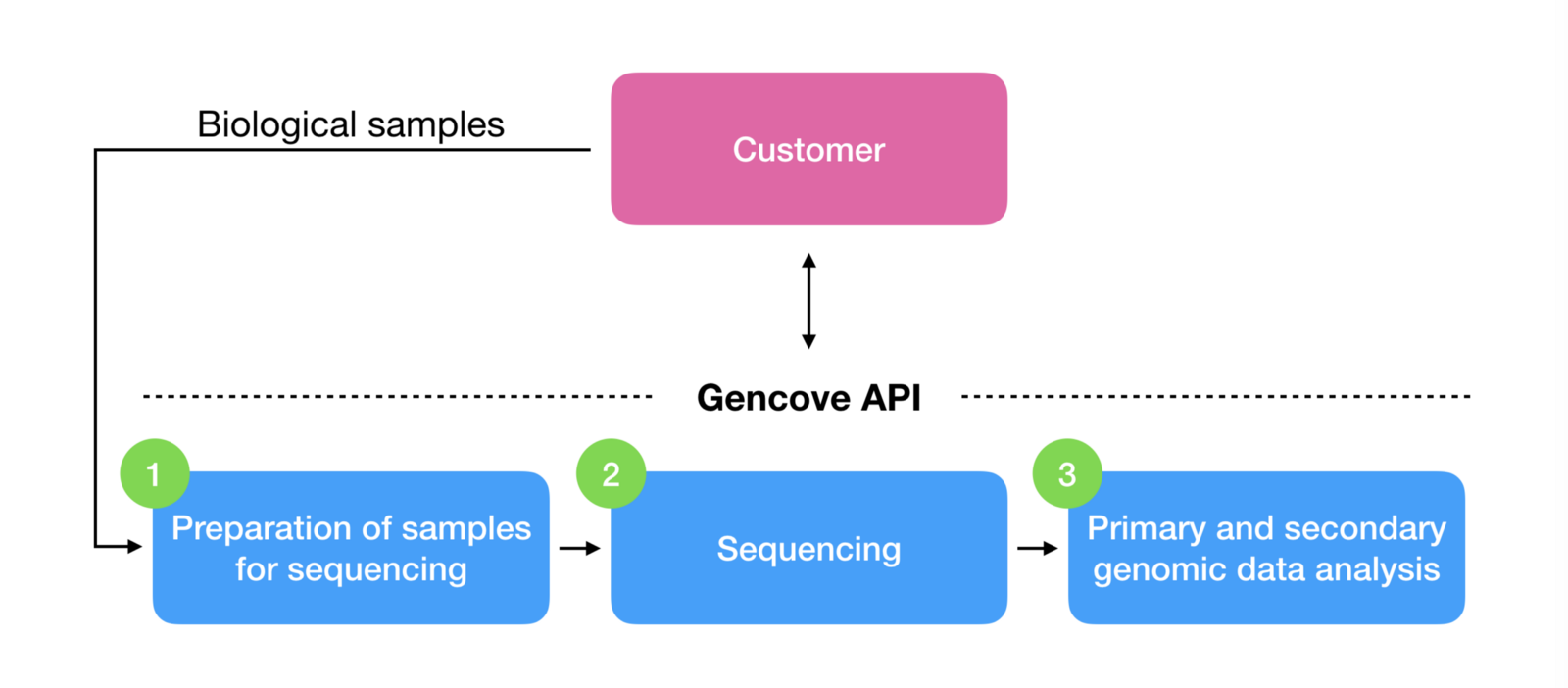
For customers with special requirements, we also happily provide our technology as an easy-to-use lab kit for processing biological samples on-premises and expose our standard data analysis pipeline as-a-service.
The technology has proven to be a great fit for population-scale studies done by pharmaceutical companies, biotechs, large academic projects, and biobanks, such as GlaxoSmithKline, Kallyope, Harvard University, Stanford University, and many others.
In this blog post, I’ll describe one of the challenges we’ve overcome on the computational side — scaling our data analysis workflow.
The Terraform infrastructure template and sample Python code for the architecture outlined in this post are freely available here under the MIT license.
Genomic data analysis
Typical analysis of genomic data consists of multiple interdependent Extract Transform and Load (ETL) processes that transform raw data from sequencing machines into more meaningful downstream data used by researchers and clinicians.
Most commonly, these processes are Docker containers running as jobs (with diverse CPU, memory, and storage requirements) on high-performance computing (HPC) clusters.
Reference architecture
Thankfully, the AWS team provides a reference architecture for building these analysis pipelines on top of AWS services like Batch, Step Functions, Lambda, DynamoDB, CloudWatch, etc.
To summarize this architecture, one AWS Step Function execution is used per sample to orchestrate the high-level workflow by scheduling and monitoring the required jobs on an HPC cluster managed by AWS Batch.
Scalability constraints
The architecture set out above worked great, but due to the specific nature of our work, we started hitting a few scalability constraints.
Since low-pass sequencing is the core technology at Gencove and the newest sequencing machines produce vast amounts of data, a state-of-the-art Illumina NovaSeq sequencer will yield raw data for 1200–1800 samples (depending on project specifics).
A quick look at the workflow layer in the reference architecture reveals the job status poller pattern, which submits a job and repeatedly polls the Batch API for job status (see figure below).
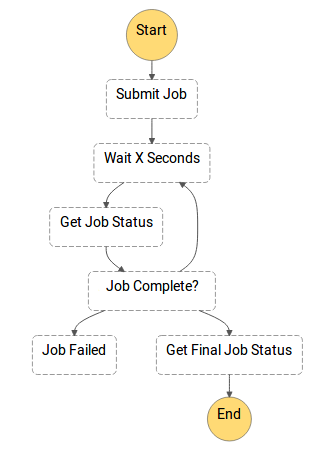 Starting 1500 state machines in parallel (one per sample), each with multiple SubmitJob API calls, will exhaust the Batch API which will start returning HTTP 429 Too Many Requests errors. Even spacing out the Step Function executions in time does not help, since subsequent DescribeJobs API calls in the job status poller keep hammering away at the API.
Starting 1500 state machines in parallel (one per sample), each with multiple SubmitJob API calls, will exhaust the Batch API which will start returning HTTP 429 Too Many Requests errors. Even spacing out the Step Function executions in time does not help, since subsequent DescribeJobs API calls in the job status poller keep hammering away at the API.
Decreasing the polling frequency, combining jobs into array jobs, combining job status queries into a single API call, and additional exponential backoffs did little to alleviate the issue.
To summarize, the issues outlined above (and their solutions) can be divided into two main categories:
1) SubmitJob API calls need to be rate-limited
This can be solved by placing SubmitJob requests on a queue and rate-limiting the consumer(s) of that queue.
2) Repeated DescribeJobs API calls need to be rate-limited or completely avoided if possible
Luckily, Step Functions allow offloading long-running tasks as “Activities” and Batch sends job state changes to CloudWatch events, which makes this category much easier to tackle by using an event-driven approach rather than rate-limited polling.
Updated architecture
Instead of using a separate queue to rate-limit SubmitJob API calls, we use the Step Function Activity as a queue for rate-limiting SubmitJob API calls in addition to using it for offloading long-running tasks in Step Functions.
An overview of the solution is provided in the figure below.
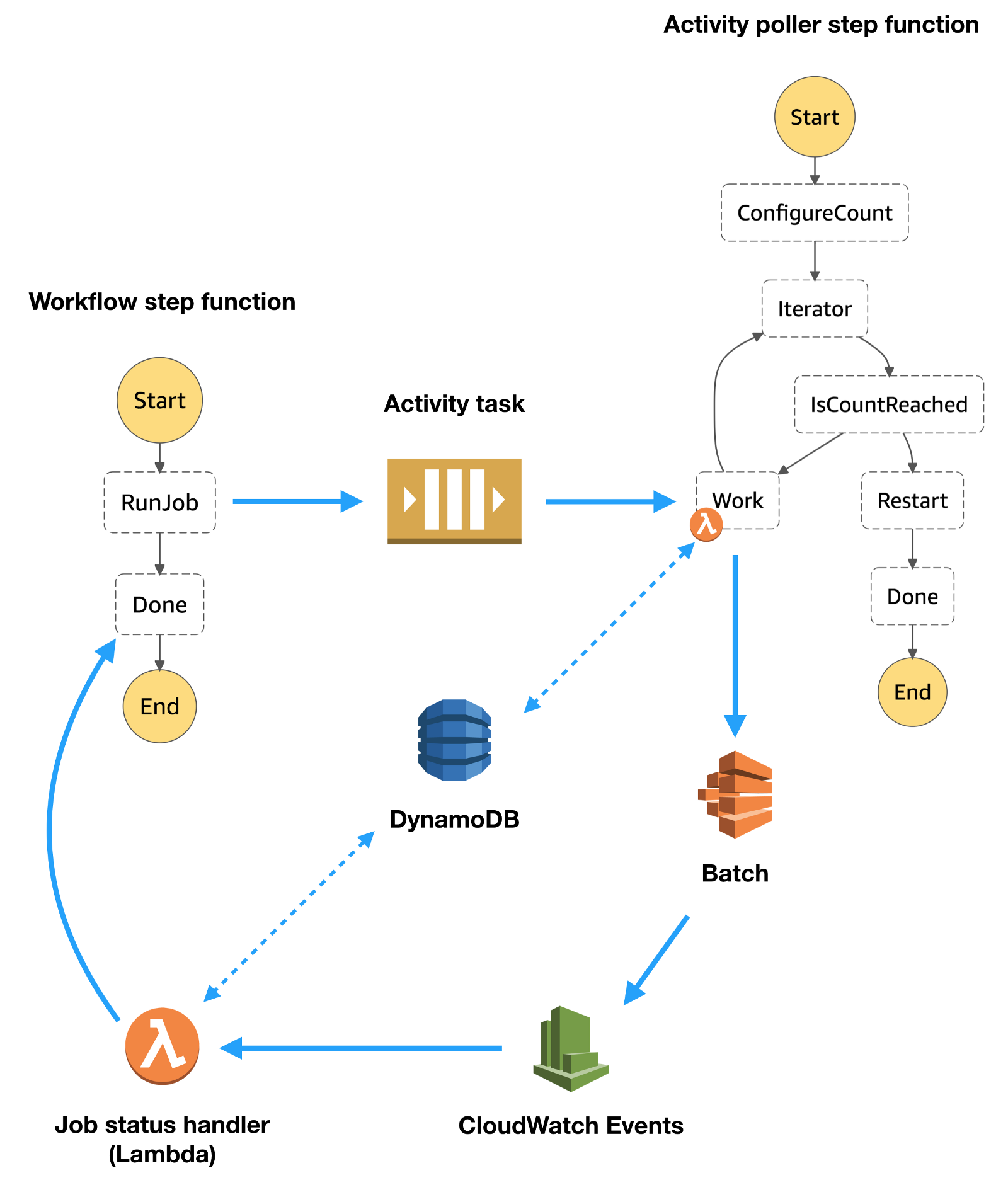
Workflow step function
- Start a Step Function Activity Task and wait for its completion
- Continue to next step in workflow
In addition to avoiding continuously polling the Batch API, using Activity Tasks avoids hitting the “Maximum Execution History Size” limit in Step Functions for long-running jobs when using the job status poller pattern.
Activity poller step function
We’ve implemented long-polling the Step Function Activity as another Step Function modeled on the Continue as a New Execution pattern. This pattern runs a Lambda function for a predefined number of times and respawns itself in order to avoid hitting the “Maximum Execution History Size” limit in Step Functions. An overview of the activity poller is:
- Long-poll the Step Function Activity to acquire the task token
- Schedule Batch job(s) with the task token in the Batch job parameters
- Update the DynamoDB item with the scheduled job id(s), using the task token as the partition key
- Go to #1 (or start a new activity poller and exit).
Running one execution of this Step Function effectively serializes SubmitJob API calls, but there is no reason multiple executions cannot run in parallel as long as the rate of SubmitJob API calls is monitored to avoid hitting Batch API limits (HTTP 429 Too Many Requests).
Job status handler
This is simply a Lambda triggered by Batch job state changes, which can be configured to fire only on transitions to SUCCEEDED or FAILED states as outlined here.
Once triggered, it:
- Notes the state update(s) in DynamoDB using the task token (stored in Batch parameters)
- If all required jobs have been noted in the DynamoDB item as completed, unblock the Activity Task using the task token
If the workflow utilizes large array jobs, handling only the parent job is often sufficient and storing state for every child job may not always be necessary.
Final thoughts and future directions
The above updates have enabled us to process thousands of samples in parallel and our only scalability limitations have been submitting service limit increase requests from time to time.
In addition to using Step Functions for workflow management, Nextflow is a great choice for workflow management on top of AWS Batch and we are keen to find a place for it in our production environment.
Sound fun? Join us!
At Gencove, we develop software and algorithms for processing genomic data at scale. We build using Python/C++ and continuously deploy to the AWS ecosystem with the help of Git, Docker, and Terraform. If that sounds fun, consider joining our team!
Acknowledgments
Thanks to Angel Pizarro, Paul Underwood, and Alexis Moinpour from the AWS team for their great advice and feedback ?
Footnotes
For a great high-level explanation of imputation, see this blog post from our friends at DNA.land
More about the performance of low-pass sequencing is available here, here, and here.