AWS for Industries
Build a medical image analysis pipeline on Amazon SageMaker using the MONAI framework
Medical imaging has transformed healthcare in a way that allows clinicians to identify diseases, make better diagnosis, and dramatically improve patient outcomes. Digital medical images have several modalities including ultrasound, X-ray, computed tomography (CT) scans and magnetic-resonance imaging (MRI) scans, positron emission tomography (PET) scans, retinal photography, histology slides, and dermoscopy images. Recent developments in deep learning algorithms, such as connected convolutional networks, are helping to identify and classify patterns in medical images that can supplement diagnostics and decision-making to offer clinicians insights into diseases, reduce costs and physician burn-out by automating repetitive tasks, and enable quicker triaging of patients with high-risk conditions.
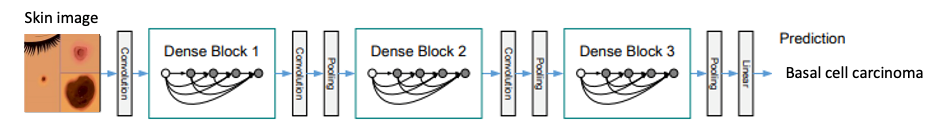 Depiction of Densely Connected Convolutional Network Model to Classify Skin Cancer
Depiction of Densely Connected Convolutional Network Model to Classify Skin Cancer
Deep learning in medical image analysis has unique challenges and it requires approaches specific to the domain to improve model performance. For example, deep learning requires large annotated datasets to train the models, but there are limited public datasets available for medical imaging. This necessitates deep learning techniques to deal with less data and imbalanced datasets where there are disproportionate observations. Medical images also have high resolution and require large amounts of storage and compute for training and hosting of models, consequently architectures are needed to address those infrastructure requirements while optimizing costs. In addition, there are very few standards or consistency on the software toolkits (ex. NiftyNet, DeepNeuro, Clara Train) used for deep learning in healthcare imaging research. This makes it difficult for researchers to collaborate on defining best practices and reproducing research.
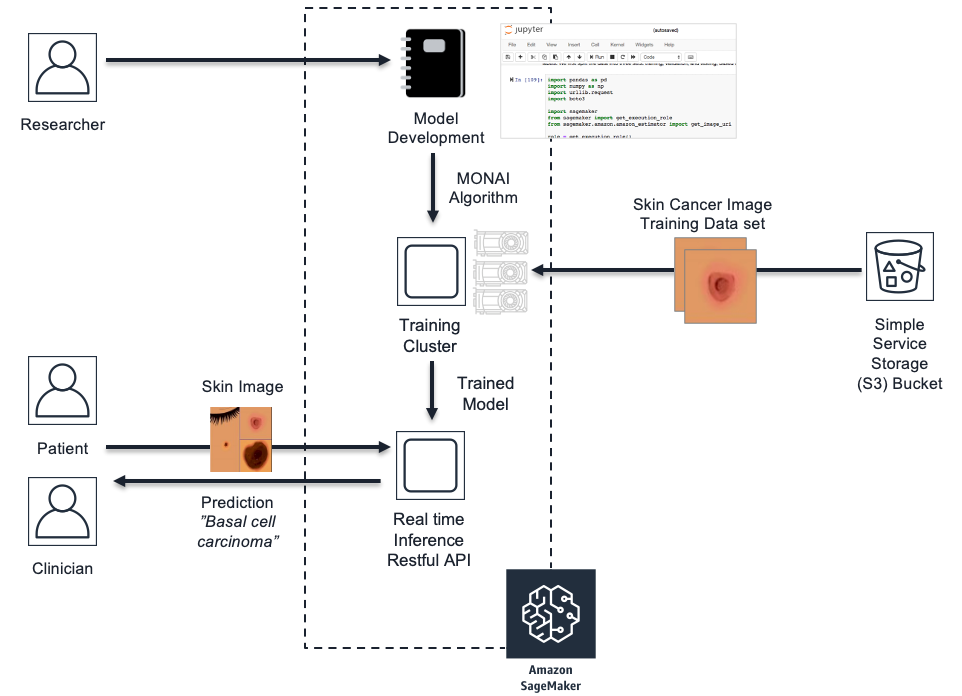
In 2018, AWS released the Registry of Open Data, which allows researchers to publish and share public datasets. AWS also released Amazon SageMaker Ground Truth, which has data labeling workflows to assist researchers in building highly accurate training sets. Ground Truth is one component of the Amazon SageMaker stack which is a fully managed service that allows researchers to build, train, and deploy their machine learning models in a single interface without having to manage servers. SageMaker provides common machine learning algorithms that are optimized to run against extremely large data in a distributed environment, and has built-in native support for bring-your-own algorithms and open source frameworks to enable interoperability. SageMaker allows researchers to train and deploy models easily in a highly scalable environment while only being charged for what they use.
Medical Open Network for AI (MONAI) was released in April 2020 and is a PyTorch-based open-source framework for deep learning in healthcare imaging. As stated on their website, project MONAI is an initiative “to establish an inclusive community of AI researchers for the development and exchange of best practices for AI in healthcare imaging across academia and enterprise researchers.” MONAI features include flexible pre-processing transforms for multi-dimensional data, portable API(s) for ease of integration into existing workflows, and domain specific implementations for networks, losses, and evaluation metrics.
In this post, we will demonstrate how to integrate the MONAI framework into the SageMaker managed service. We will also give example code of MONAI pre-processing transforms that can assist with imbalanced datasets and image transformations. Then, we will review the code to invoke MONAI neural network architectures such as Densenet for image classification and explore structure of PyTorch code to train and serve the model within SageMaker. Additionally, we will cover the SageMaker API calls to launch and manage the compute infrastructure for both model training and hosting for inference. The examples are based on a skin cancer classification model that predicts skin cancer classes and uses the HAM10000 dermatoscopy skin cancer image dataset published by Harvard.
Prerequisites
We shared the sample model code in the GitHub repo. If you would like to deploy the model in your account, make sure you have the following in place:
- An AWS account that you have administrative permissions
- GitHub account
- Prior experience with Jupyter Notebook and Python is advised
Build MONAI models in SageMaker
SageMaker provides tools that researchers are familiar with, like managed Jupyter Notebooks and Docker containers. You can build your model from built-in algorithms or bring-your-own algorithm using several options such as script mode using built-in containers, deploy your own container, or use a container from the AWS Marketplace.
For the skin cancer model, we will use script mode by leveraging an existing PyTorch built-in container and extend it with the MONAI package for our custom algorithm. In script mode, you create a training script in a separate file from the Jupyter Notebook that you will use to launch the model training job. In our skin cancer example, we have created a script called monai_skin_cancer.py.
In the same directory as your training script, create a requirements.txt file to include MONAI and any additional packages that you will need installed in the PyTorch built-in container to support the training.
Example: requirements.txt
The training script has access to several properties about the SageMaker training environment through environment variables, such as number of GPUs available to host, hyperparameters, and directory for model artifacts. The training code should be written after the main guard (if __name__ == '__main__':) similar to the following example that calls the train function for model training.
Example monai_skin_cancer.py: PyTorch training script entry point
The train function will ingest the skin cancer training dataset using MONAI transforms for pre-processing and augmentation. MONAI has transforms that support both Dictionary and Array formats, specialized for the high-dimensionality of medical images. The transforms include several categories such as Crop & Pad, Intensity, IO, Post-processing, Spatial, and Utilities. In the following excerpt, the Compose class chains a series of image transforms together and returns a single tensor of the image.
Example monai_skin_cancer.py: MONAI image transforms
MONAI includes deep neural networks such as UNet, DenseNet, GAN, and others and provides sliding window inferences for large medical image volumes. In the skin cancer image classification model, we train the MONAI DenseNet model on the skin cancer images loaded and transformed using the DataLoader class in previous steps for thirty epochs while measuring loss.
Example monai_skin_cancer.py: Train MONAI DenseNet model
To deploy your trained MONAI model to SageMaker in later steps, you must save the model defined by environment variable SM_MODEL_DIR in your training script.
Example monai_skin_cancer.py: Save the model
Once your training job is complete, SageMaker compresses and uploads the trained model to the Amazon Simple Storage Service (S3) bucket that you specified in your job. We will review how to create the training job and deploy your model for inference in the next sections.
SageMaker Life Cycle for Skin Cancer Model
Scale up training of MONAI models
SageMaker uses ephemeral clusters for training each model. Ephemeral clusters are dedicated instances that are alive for the number of seconds that the training job is running. This lets you build the model within your managed Jupyter Notebook on an instance and train the model on independent instances to avoid conflicting package installations and resource dependencies and optimize costs.
The following example shows how to use the SageMaker Python SDK in a Jupyter Notebook to instantiate a training job and execute the skin cancer model training script that we discussed in previous sections.
Example monai_skin_cancer_notebook.ipynb: Training job to execute training script
When you call the fit method in this example, it will launch the training instance with a compute size based on the train_instance_type parameter, instantiate the built-in PyTorch deep learning container, install the MONAI dependent packages found in requirements.txt, download the skin cancer training dataset from Amazon S3, and execute the training script referenced by the entry_point parameter. Given the small size of the skin cancer training dataset, we were able to download it to the training instance Amazon Elastic Block Store (EBS) volume. But in the case of large medical image datasets, you can use SageMaker Pipe input mode to stream the data directly to your training instances. For more details, see Pipe input model for Amazon SageMaker algorithms.
SageMaker also supports multi-machine distributed training using the PyTorch Estimator for training models on large datasets. If you specify train_instance_count greater than one, SageMaker will launch multiple instances for a training cluster and execute the training script on each host. The design patterns and implementation details of this approach are addressed in Scalable multi-node deep learning training using GPUs in the AWS Cloud.
SageMaker has monitoring metrics for training jobs such as memory, disk, GPU utilization, training error, and others that gives visibility into model training. MONAI provides evaluation metrics focused on the quality of medical image models such as Mean Dice for image segmentation. The integration of SageMaker and MONAI allows researchers to get a comprehensive view into their medical imaging model training.
In the skin cancer model training, we used the MONAI method compute_roc_auc to measure the area under the ROC (receiver operating characteristic) curve.
Example monai_skin_cancer.py: Area under the ROC curve
Deploy MONAI model on RESTful API Endpoint
SageMaker hosts models via model endpoints that are composed of one or more managed Amazon Elastic Compute (EC2) instances which are deployed over different availability zones to increase the availability of your inferencing services. Each Amazon EC2 instance contains a web server to respond to requests and your model artifacts so that each instance can serve prediction responses. The health of those instances is maintained with a load balancer which can distribute the requests, and auto scaling which can launch new instances during peak loads. The model endpoint is a RESTful API which SageMaker creates for your specific model when you call estimator.deploy.
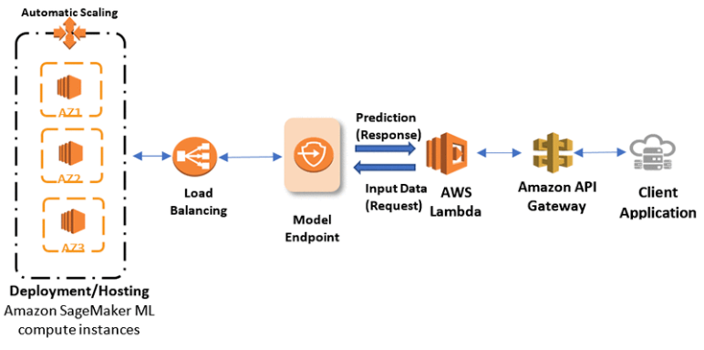 SageMaker Model Endpoint with AWS Lambda as Client
SageMaker Model Endpoint with AWS Lambda as Client
Example monai_skin_cancer_notebook.ipynb: Deploy the model to managed endpoint
The endpoint you create runs a PyTorch model server on the instance that loads the trained model that was saved by your training script. The server loads your model by invoking model_fn, which we included in the training script for the MONAI skin cancer model.
Example monai_skin_cancer.py: Load the MONAI skin cancer model
Once SageMaker has loaded your trained model, it will serve the model by responding to inference requests when you call predictor.predict. In the skin cancer model, we submitted a patient skin cancer image and the model predicted the class and probability.
Example monai_skin_cancer_notebook.ipynb: Predict the skin cancer class and show probability
Given that each skin cancer image in the test dataset for our example was less than two megabytes in size, we were able to use the real-time RESTful API endpoint. Use cases that have large medical image sizes or large volumes of images to infer should consider using batch transform for inferences. The batch transform solution is explored in Performing batch inference with TensorFlow Serving in Amazon SageMaker.
Conclusion
The integration of SageMaker and MONAI lets researchers define best practices in medical image analysis and collaborate on research using MONAI as a common foundation, while leveraging familiar tools provided in SageMaker on a managed environment that is scalable, secure, and cost effective. In this post, we explored some foundational features of MONAI in SageMaker. In future research, there are opportunities in supervised training engines, federated learning, and event handling for end-to-end workflows.
To learn more about how AWS is accelerating innovation in healthcare, please visit aws.amazon.com/health.
References
[1] Shen, Dinggang et al. “Deep Learning in Medical Image Analysis.” Annual review of biomedical engineering vol. 19 (2017): 221-248. doi:10.1146/annurev-bioeng-071516-044442
[2] Tschandl, P., Rosendahl, C. & Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5, 180161 (2018). https://doi.org/10.1038/sdata.2018.161
[3] MONAI, Medical Open Network for AI monai.io