Artificial Intelligence
Accelerating genomics variant interpretation with AWS HealthOmics and Amazon Bedrock AgentCore
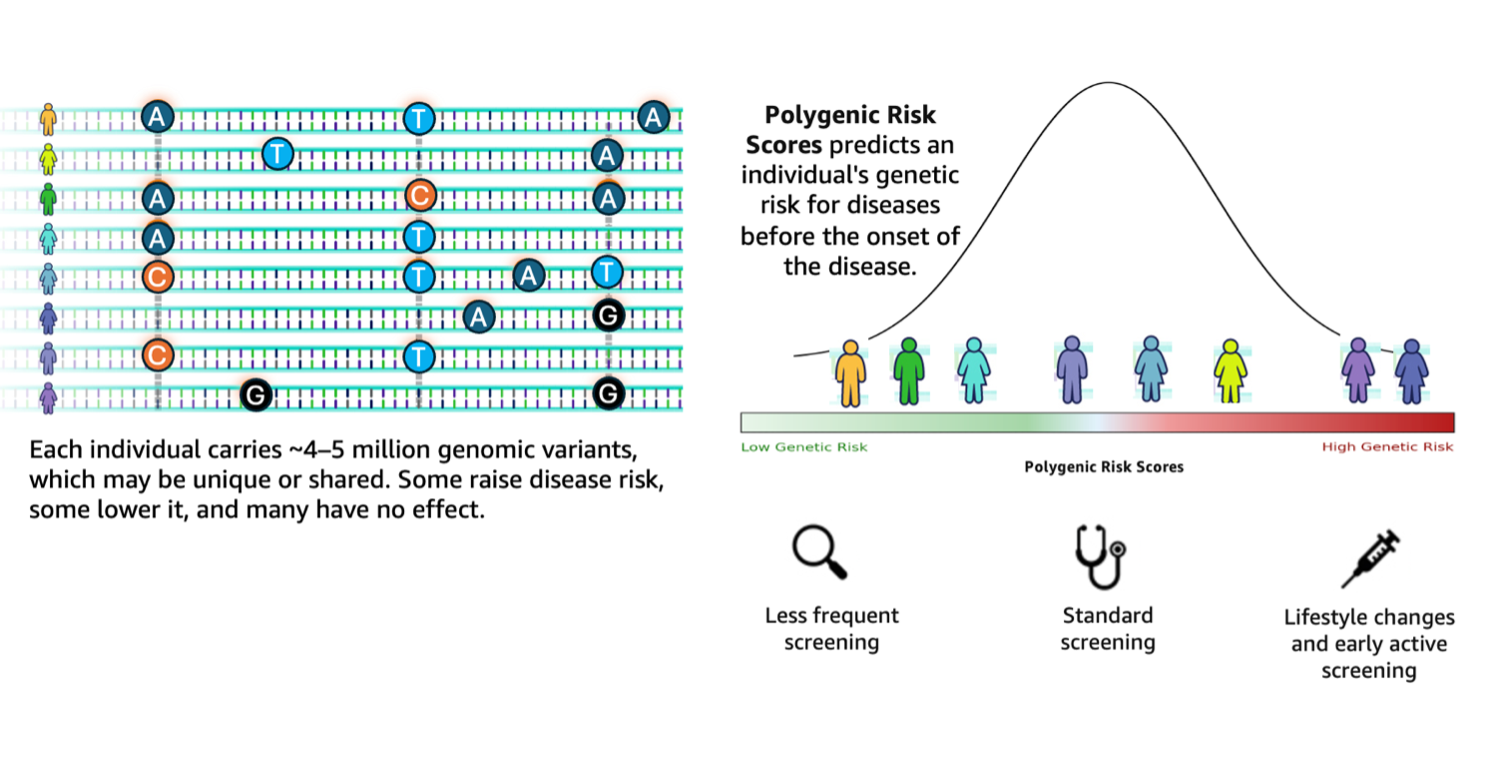
Genomic research stands at a transformative crossroads where the exponential growth of sequencing data demands equally sophisticated analytical capabilities. According to the 1000 Genomes Project, a typical human genome differs from the reference at 4.1–5.0 million sites, with most variants being SNPs and short indels. These variants, when aggregated across individuals, contribute to differences in disease susceptibility captured through polygenic risk scores (PRS). Genomic analysis workflows struggle to translate such large-scale variant data into actionable insights. They remain fragmented, requiring researchers to manually orchestrate complex pipelines involving variant annotation, quality filtering, and integration with external databases such as ClinVar.
AWS HealthOmics workflows along with Amazon S3 tables and Amazon Bedrock AgentCore together provide a transformative solution to these challenges. HealthOmics workflows support the seamless integration of annotating Variant Call Format (VCF) files with insightful ontologies. Subsequently, the VEP-annotated VCF files need to be transformed into structured datasets stored in optimized S3 tables to improve query performance across large variant cohorts. The Strands Agents SDK running on Amazon Bedrock AgentCore provides a secure and scalable AI agent application so that researchers can interact with complex genomic datasets without specialized query expertise.
In this blog post, we show you how agentic workflows can accelerate the processing and interpretation of genomics pipelines at scale with a natural language interface. We demonstrate a comprehensive genomic variant interpreter agent that combines automated data processing with intelligent analysis to address the entire workflow from raw VCF file ingestion to conversational query interfaces. Most importantly, this solution removes the technical expertise barrier that has traditionally limited genomic analysis to specialized bioinformaticians. This enables clinical researchers to upload raw VCF files and immediately ask questions like ‘Which patients have pathogenic variants in BRCA1?’ or ‘Show me drug resistance variants in this cohort’. The code for this solution is available in the open-source toolkit repository of starter agents for life sciences on AWS.
Understanding variant annotation in genomic analysis
The foundation of genomic variant interpretation relies on comprehensive annotation pipelines that connect raw genetic variants to biological and clinical context. Variant Effect Predictor (VEP) and ClinVar represent two essential components in modern genomic analysis workflows, each providing complementary information that researchers must integrate to derive meaningful insights.
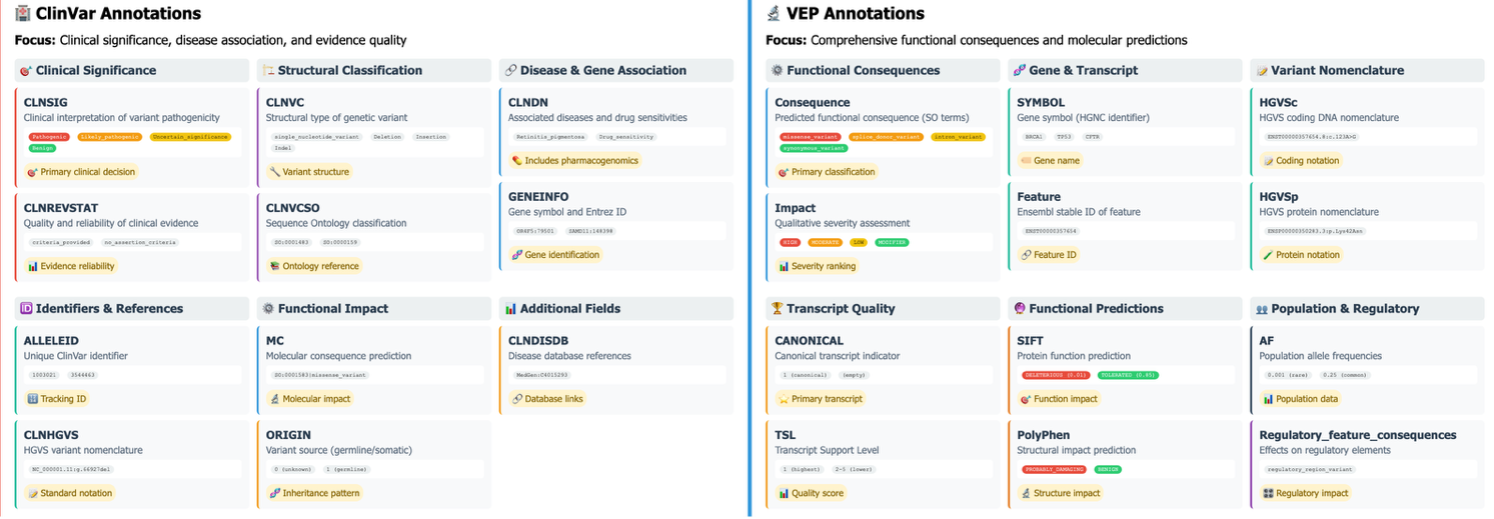
The comparative visualization illustrates the distinct yet complementary annotation capabilities of ClinVar and VEP for genomic variant interpretation. ClinVar annotations (left) focus primarily on clinical significance assessment, providing curated pathogenicity classifications (CLNSIG), evidence quality metrics (CLNREVSTAT), and disease associations (CLNDN) directly relevant to clinical decision-making. VEP annotations (right) deliver comprehensive functional information including consequence types (missense_variant, synonymous_variant, intron_variant), impact severity classifications (HIGH, MODERATE, LOW, MODIFIER), gene symbols, and transcript-specific effects with detailed positional information.
Current annotation workflow challenges
Variant annotation workflows typically follow a sequential process that includes:
- Initial VCF processing: Raw variant call format (VCF) files from sequencing systems require preprocessing to normalize representation and filter low-quality calls.
- VEP annotation: Running the Variant Effect Predictor tool requires substantial computational resources, especially for whole genome sequencing data with millions of variants per sample. VEP analysis can take 2-8 hours for a single genome depending on available compute resources and annotation depth.
- ClinVar integration: Clinical annotations must be retrieved from ClinVar and matched to variants through a separate process, requiring database lookups and format conversions.
- Multi-sample integration: Creating cohort-level analyses requires complex joining operations across samples, typically performed with specialized tools that generate large, flat files difficult to query efficiently.
- Interpretation: Scientists must then use various tools to filter, sort, and analyze the annotated data—a process that often requires custom scripts and significant bioinformatics expertise. This technical bottleneck means that clinical researchers cannot independently explore their genomic data, creating delays of days or weeks between asking a biological question and receiving an answer.
Dataset complexity and scale
The scale of genomic variant analysis is exemplified by datasets like the 1000 Genomes Phase 3 Reanalysis with DRAGEN, which contains:
- Over 2,500 individual samples from diverse populations
- Approximately 85 million unique variants across all samples
- Multiple annotation versions (DRAGEN 3.5, 3.7, 4.0, and 4.2) that must be reconciled
- Complex structural variants alongside SNPs and indels
This complexity creates significant bottlenecks in traditional analysis pipelines that rely on flat file processing and manual integration steps.
Solution overview
Building genomic cohorts or computing PRS across multiple patients demands significant compute resources to generate joint variant call tables and comprehensive annotations using tools like the Variant Effect Predictor (VEP). Most critically, these workflows create a technical barrier where only bioinformaticians with SQL expertise and deep understanding of variant file formats can extract meaningful insights, leaving clinical researchers dependent on specialized technical teams for basic genomic queries.
The transformative advantage of our AI-powered approach lies in democratizing genomic analysis through natural language interaction. While traditional VEP pipelines require days of technical expertise to answer clinical questions like ‘Which patients have high-impact variants in drug resistance genes?’, with our solution researchers can ask these questions conversationally and receive answers in minutes. This represents a shift from technical dependency to self-service genomic insights so that clinical researchers, tumor boards, and genomics teams to directly explore their data without waiting for bioinformatics support.
Our solution demonstrates a generative AI-powered genomics variant interpreter agent that combines automated data processing with intelligent natural language analysis. The architecture addresses the entire genomic analysis workflow, from raw VCF file ingestion to conversational query interfaces.
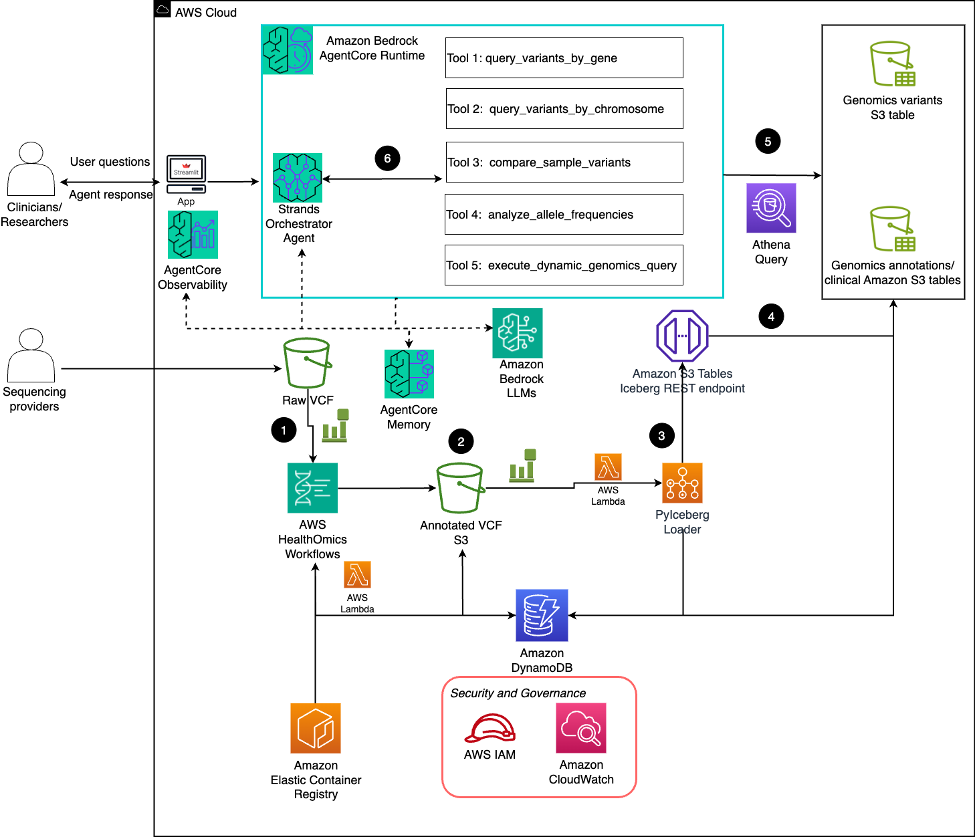
The solution follows six key steps that transform raw genomic data into actionable insights:
- Raw VCF processing: Raw VCF files from sequencing providers are uploaded to Amazon S3 storage and trigger AWS Lambda functions through S3 event notifications, which orchestrate AWS HealthOmics workflows.
- VEP annotation: AWS HealthOmics workflows automatically process raw VCF files using the Variant Effect Predictor (VEP), enriching variants with functional predictions and clinical annotations in parallel before storing the annotated results back to S3.
- Event coordination: Amazon EventBridge monitors workflow completion and triggers Lambda functions that update job status in Amazon DynamoDB and AWS Batch Fargate compute environment transforms VEP annotated VCF files and ClinVar annotations into Iceberg format as PyIceberg module
- Data organization: PyIceberg loader interacts with the Amazon S3 Tables Iceberg Rest Endpoint. Amazon S3 Tables connects registers the table metadata in AWS Glue Data Catalog. Schema information (columns, data types, partitions) gets catalogued for annotated VCF and ClinVar annotations. It also establishes analytics connector for downstream analytics.
- SQL-powered analysis: Amazon Athena provides SQL-based querying capabilities over the genomic data through columnar storage format, enabling large-scale analysis with ideal query responses across millions of variants.
- Natural language interaction: The Strands orchestrator agent, powered by Amazon Bedrock LLMs on AgentCore Runtime, provides a natural language interface through five specialized tools that execute Athena queries:
- query_variants_by_gene: Retrieves variants associated with specific genes
- query_variants_by_chromosome: Facilitates chromosome-specific variant analysis
- compare_sample_variants: Enables comparative genomics across patient samples
- analyze_allele_frequencies: Provides population genetics insights
- execute_dynamic_genomics_query: Supports flexible, ad-hoc analysis requests
The architecture includes comprehensive security controls through AWS IAM for fine-grained access management and Amazon CloudWatch for monitoring. The automated, event-driven pipeline supports scalable parallel processing of VCF files that automatically adapts to growing genomic datasets while maintaining consistent annotation quality and analytical capabilities.
Amazon S3 Tables with PyIceberg: Transforming VCF to a structured cohort
Amazon S3 Tables with PyIceberg transforms VEP-annotated VCF files into a structured cohort, queryable datasets optimized for AI-driven analysis. This creates the data foundation for natural language interfaces to efficiently interact with complex genomic data.
PyIceberg creates Apache Iceberg tables in S3 Tables format, provide the following benefits:
- Optimal queries: The agent can perform complex genomic queries across millions of variants with minimal latency through optimized columnar storage, transforming analyses that previously required hours of SQL development and execution into instant conversational responses.
- Rich annotation access: The VEP and ClinVar annotations become directly queryable through SQL via Amazon Athena, allowing the AI agent to extract specific genomic insights
- Cohort-level analysis: The structured Iceberg format (PyIceberg) supports efficient comparisons across patient cohorts for population-level queries through natural language.
The separation of variant data from annotation data in S3 Tables creates an ideal foundation for AI-driven analytics because genomics variants S3 tables contain core positional information that agents can rapidly filter, and the annotations/clinical S3 tables house the rich functional and clinical context needed for interpretation. With this structure, the Strands agent can construct targeted queries that precisely answer user questions through the AWS Glue Data Catalog Connector.
This conversion from raw VCF files to structured tables is what makes it possible for researchers to query complex genomic datasets conversationally through the Strands orchestrator agent [KM1] on Amazon Bedrock AgentCore.
Intelligent genomic analysis with Strands Agents and AgentCore Runtime
The conversational interface represents the core innovation of our genomics AI solution, built using the Strands Agents SDK and deployed on Amazon Bedrock AgentCore Runtime. This sophisticated AI agent understands complex genomic concepts and translates natural language queries into appropriate analytical operations against the structured genomic datasets.
AgentCore Runtime is a secure, serverless runtime purpose-built for deploying and scaling dynamic AI agents and tools. This solution offers several key advantages for genomic analysis:
- Model and framework flexibility: AgentCore services are composable and work with open source or custom framework and models, both in and outside of Amazon Bedrock
- Multi-hour agentic workloads: Supports long-running workloads up to 8 hours and payloads up to 100MB
- Security: Dedicated microVMs for each user session with complete isolation
- Enterprise-grade integration: Built-in authentication via AgentCore Identity with AWS IAM
- Observability: Comprehensive tracing of agent reasoning and tool invocations
- Private resource access: Connectivity to databases and APIs within Amazon Virtual Private Cloud
- Faster time-to-market: Accelerated deployment and development cycles for AI agent solutions
For detailed information on Amazon Bedrock AgentCore capabilities, refer to the Amazon Bedrock AgentCore documentation.
Strands Agents provide a robust foundation for building domain-specific AI agents with specialized capabilities through a model-driven approach that orchestrates genomic analysis tools using an agentic loop concept. This iterative reasoning framework enables agents to dynamically select and execute appropriate tools based on analysis requirements. Our genomic variant interpreter implements five key tools that leverage the structured data created by Amazon S3 Tables:
- Variant querying: Translates gene-based questions into precise Athena SQL queries that retrieve associated variants.
- Chromosome analysis: Enables region-specific genomic interrogation through natural language.
- Sample comparison: Facilitates cross-patient genomic analysis without requiring SQL joins.
- Population frequency analysis: Contextualizes findings against reference datasets like 1000 Genomes.
- Dynamic query generation: Converts complex natural language requests into optimized SQL.
Natural language queries
The agent demonstrates remarkable capability in handling diverse query types. In the traditional model clinical researchers must wait for bioinformatics teams to write custom scripts and run complex analyses. Instead of spending days crafting SQL queries and wrestling with VCF file formats, researchers can now explore their genomic data as naturally as having a conversation with a genomics expert.
Cohort-level analysis
User: "Summarize as a table the total number of variants and pathogenicity per patient in this cohort?"
For this query, the agent:
- Uses the execute_dynamic_genomics_query tool.
- Analyzes variant data across the cohort of samples.
- Generates a comprehensive cohort summary with patient counts and variant statistics.
- Presents findings in a structured and tabular format summary.
Cohort-level frequency analysis
User: "Provide me the allelic frequencies of shared pathogenic or likely pathogenic variants in this cohort and 1000 genomes?"
The agent translates this into queries that:
- Retrieve the list of pathogenic variants for the patient by running the execute_dynamic_genomics_query and analyze_allele_frequencies tool.
- Filter for clinically relevant pathogenic variants.
- Extract disease level information from ClinVar and allele frequencies from VEP.
- Present results with relevant context.
Comorbidity risk association
User: " Which are those patients have variant in ADRA2A gene at chr10:111079820 and, does these patients have any additional high impact variants linked with statin or insulin resistance? "
For this query, the agent:
- Searches for additional risk variants in drug resistance pathways for a specific disease context.
- Connect with clinical significance at individual patient level for comorbidity.
- Provide clinical implications of joint clinical and drug resistance pathways.
This natural language interface minimizes the need for researchers to master complex SQL syntax or understand the underlying data structures, democratizing access to genomic insights across clinical and research teams regardless of their technical background.
Advanced analytic processing
In addition to queries, the genomics variant interpreter agent demonstrates advanced analytical capabilities that extend beyond basic variant identification. Researchers can explore complex questions that traditionally required days of analysis.
Clinical decision support
User: " Perform a thorough analysis on patient NA21144 and provide me the risk stratification for this patient"
For this query, the agent:
- Analyzes variants in disease pathways genes, pharmacogenomics, and provides evidence-based recommendations.
- Performs risk stratification by combining variant impact predictions with clinical significance classifications.
- Identifies variants of uncertain significance.
- Flags high-impact variants in clinically relevant genes.
Pharmacogenomics guided-dosing strategy
Researchers can leverage the agent for sophisticated pharmacogenomics pathway analyses across large cohorts through queries like:
User: " Which major drug-related pathways are significantly enriched with genetic variants in this patient cohort? Provide me the most impactful pharmacogenomic pathways and associated patient IDs "
This allows exploration of variant frequency distributions, consequence type patterns, and gene-level variant burdens across different populations—all through conversational interfaces without complex SQL or bioinformatics pipelines.
Benefits and limitation
The solution helps to solve the current challenges:
| Challenges | Solutions |
| Initial VCF processing – Low-quality calls | The agent automatically prechecks quality calls of variants before making variant interpretation decisions |
| VEP annotation at scale | The solution automates VCF annotation at scale of 20 in batches uses right compute resource to achieve the appropriate performance. |
| ClinVar integration | The agent assess the query context and joint-query will be built dynamically based on the user interest. |
| Multi-sample integration | Amazon S3 Tables integration in Iceberg format makes the cohort of VCF files to query with ideal performance. |
| Genomics interpretation | The agent understands the context and user interest to make the informed decisions carefully reason out based on the appropriate evidences from the annotations and inhouse. |
The solution has the following limitations:
- Lambda Runtime constraints: The current implementation uses AWS Lambda for VCF/GVCF processing, which has a maximum execution time of 15 minutes. This constraint may be insufficient for loading large VCF files or especially large GVCF files into Iceberg S3 Tables, as these operations can take substantially longer than the Lambda timeout limit. For production workloads with large genomic datasets, consider using AWS HealthOmics workflows, AWS Batch, ECS tasks, or EC2 instances with longer execution times to handle the data loading process.
- Schema optimization trade-offs: The schema implementation uses sample and chromosome partitioning, which is optimized for patient-level analysis. However, cohort-level analysis typically requires different partitioning strategies and schema designs to achieve optimal performance at scale. Making both patient-level and cohort-level analytics performant within a single schema becomes increasingly challenging as cohort sizes grow beyond hundreds of samples. For large-scale cohort studies (thousands to tens of thousands of samples), consider implementing separate schemas or materialized views optimized for specific analytical patterns, or explore denormalized structures that better support population-level queries.
Future technological evolution
The solution’s modular architecture establishes a foundation for continued innovation in AI-powered genomic analysis. Future versions could integrate additional annotation databases, external APIs, and support multi-modal analysis combining genomic data with clinical records and imaging. Domain-specific fine-tuning on genomic data could further improve interpretation accuracy, while integration with electronic health records would provide point-of-care genomic insights.
A particularly promising direction is multi-agent collaboration in pharmaceutical R&D, where this genomics variant interpreter agent could work alongside specialized agents for drug profiling, target identification, literature evidence, and hypothesis generation. This collaborative agent framework can dramatically accelerate drug discovery pipelines by connecting variant-level insights directly to therapeutic development, streamlining the translation from genetic findings to clinical applications.
Conclusion
This next-generation genomics agentic AI solution represents a fundamental transformation in how researchers and clinicians interact with genomic data. By seamlessly integrating AWS HealthOmics for automated variant annotation and data transformation with Amazon Bedrock AgentCore for intelligent interpretation, we’ve created a comprehensive solution that addresses the entire genomic analysis workflow.
The combination of automated VEP annotation workflows, S3 Tables for transforming VCF data into queryable Iceberg tables, and Strands Agents on Amazon Bedrock AgentCore for natural language interaction creates a system that minimizes traditional barriers between variant annotation, data processing, and clinical interpretation. By automating complex technical processes and providing intuitive interaction methods, researchers can now focus on biological questions rather than technical implementation details.
As genomic data continues to grow exponentially and clinical applications become increasingly sophisticated, systems like this will become essential infrastructure for advancing precision medicine and accelerating scientific discovery. The solution demonstrated with the 1000 Genomes Phase 3 Reanalysis dataset shows how even large-scale genomic cohorts can be analyzed through simple conversational interfaces, democratizing access to advanced genomic insights.
The code for this solution is available on the Life sciences agents toolkit, and we encourage you to explore and build upon this template. For examples to get started with Amazon Bedrock AgentCore, check out the Amazon Bedrock AgentCore repository.
About the authors
 Edwin Sandanaraj is a genomics solutions architect at AWS. With a PhD in neuro-oncology and more than 20 years of experience in healthcare genomics data management and analysis, he brings a wealth of knowledge to accelerate precision genomics efforts in Asia-Pacific and Japan. He has a passionate interest in clinical genomics and multi-omics to accelerate precision care using cloud-based solutions.
Edwin Sandanaraj is a genomics solutions architect at AWS. With a PhD in neuro-oncology and more than 20 years of experience in healthcare genomics data management and analysis, he brings a wealth of knowledge to accelerate precision genomics efforts in Asia-Pacific and Japan. He has a passionate interest in clinical genomics and multi-omics to accelerate precision care using cloud-based solutions.
 Hasan Poonawala is a Senior AI/ML Solutions Architect at AWS, working with Healthcare and Life Sciences customers. Hasan helps design, deploy and scale Generative AI and Machine learning applications on AWS. He has over 15 years of combined work experience in machine learning, software development and data science on the cloud. In his spare time, Hasan loves to explore nature and spend time with friends and family.
Hasan Poonawala is a Senior AI/ML Solutions Architect at AWS, working with Healthcare and Life Sciences customers. Hasan helps design, deploy and scale Generative AI and Machine learning applications on AWS. He has over 15 years of combined work experience in machine learning, software development and data science on the cloud. In his spare time, Hasan loves to explore nature and spend time with friends and family.
 Charlie Lee is genomics industry lead for Asia-Pacific and Japan at AWS and has a PhD in computer science with a focus on bioinformatics. An industry leader with more than two decades of experience in bioinformatics, genomics, and molecular diagnostics, he is passionate about accelerating research and improving healthcare through genomics with cutting-edge sequencing technologies and cloud computing.
Charlie Lee is genomics industry lead for Asia-Pacific and Japan at AWS and has a PhD in computer science with a focus on bioinformatics. An industry leader with more than two decades of experience in bioinformatics, genomics, and molecular diagnostics, he is passionate about accelerating research and improving healthcare through genomics with cutting-edge sequencing technologies and cloud computing.