AWS HPC Blog
Tag: GPU
Introducing GPU health checks in AWS ParallelCluster 3.6
AWS ParallelCluster 3.6.0 can now detect GPU failures in HPC and AI/ML tasks. Health checks run at the start of Slurm jobs and if they fail, the job is requeued on another instance. This can increase reliability and prevent wasted spend.
Benchmarking NVIDIA Clara Parabricks Somatic Variant Calling Pipeline on AWS
Somatic variants are genetic alterations which are not inherited but acquired during one’s lifespan, for example those that are present in cancer tumors. In this post, we will demonstrate how to perform somatic variant calling from matched tumor and normal genome sequence data, as well as tumor-only whole genome and whole exome datasets using an NVIDIA GPU-accelerated Parabricks pipeline, and compare the results with baseline CPU-based workflows.
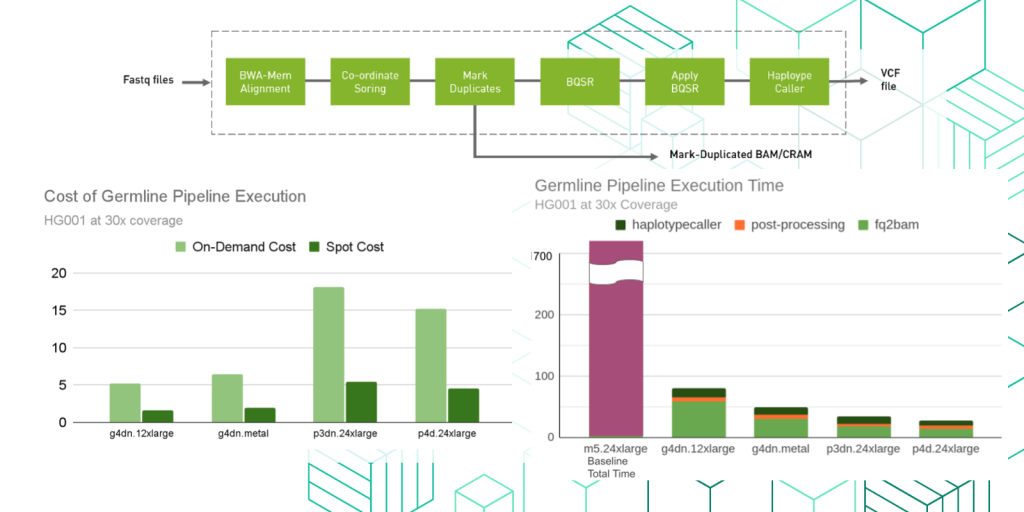
Benchmarking the NVIDIA Clara Parabricks germline pipeline on AWS
This blog provides an overview of NVIDIA’s Clara Parabricks along with a guide on how to use Parabricks within the AWS Marketplace. It focuses on germline analysis for whole genome and whole exome applications using GPU accelerated bwa-mem and GATK’s HaplotypeCaller.
Price-Performance Analysis of Amazon EC2 GPU Instance Types using NVIDIA’s GPU optimized seismic code
Seismic imaging is the process of positioning the Earth’s subsurface reflectors. It transforms the seismic data recorded in time at the Earth’s surface to an image of the Earth’s subsurface. This is done by back-propagating data from time to space in a given velocity model. Kirchhoff depth migration is a well-known technique used in geophysics for seismic imaging. Kirchhoff time and depth migration produce an image with higher resolution and generate an image of the subsurface for a subset class of the data, providing valuable information about the petrophysical properties of the rocks and helps to determine how accurate the velocity model is. This blog post looks at the price-performance characteristics computing Kirchhoff migration methods on GPUs using Nvidia’s GPU-optimized code.