AWS News Blog
Genome Engineering Applications: Early Adopters of the Cloud
Our friends at the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia sent along the guest post below to tell us about how AWS powers an important new genome editing technique.
— Jeff
Recent developments in molecular engineering technology now enables the accurate editing of genomes. The new technology, called CRISPR-Cas9, can be programmed to recognize and edit specific locations in the genome by pattern-matching unique sequences of DNA. While this is a powerful new tool for researchers, the ability to scan and identify targets across the entire genome has created unprecedented demand for large-scale computation. Earlier this year, the US National Institutes of Health (NIH) has approved the use of these technologies for human health. This has the potential to revolutionize cancer treatments and also adds a new time-critical dimension to the compute requirements.
A New Approach to Cancer Treatments
Approximately two in five people will be diagnosed with cancer at some point during their lifetime and while overall cancer survival has doubled, there are still cancer types with very low survival rate, for example just 1% for pancreatic cancer. This is mainly due to the difficulty of finding therapeutic interventions that kill cancer cells but not harm the healthy tissue in the body.
The new NIH approved trial will leverage breakthroughs in the genome editing technology, CRISPR-Cas9, to develop a different treatment approach. In this, the patient’s own immune system is boosted through specific modifications of the cells that natively fight cancer. This has the potential of being effective for a wide range of different tumors, with the current trial including patients with specific blood and solid cancers, as well as melanoma.
Cloud Services for Computationally Guided Genome Engineering
This new application in human health requires an increase in robustness and efficiency of CRISPR-Cas9 design in order to meet the time constraints of clinical care. Built on AWS cloud-services, researchers in the eHealth program of the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia, developed GT-Scan2, a novel software tool to address this issue.
“Compared to other available methods, GT-Scan2 identifies genomic location with higher sensitivity and specificity,” says Dr. Denis Bauer who is leading the transformational bioinformatics team.

GT-Scan2 shows the identified CRISPR target sites at the genomic position and annotates them with high or low activity as well as their off-target potential.
GT-Scan2 improves the effectiveness of the system by finding sites that are unique in the genome. This avoids diluting the effect due to “off-target”, which are other sites in the genome with high sequence similarity. It also optimizes robustness by finding sites that are easier to modify.
“While it was known that the three-dimensional genome organization plays a role in CRISPR binding, GT-Scan2 is the first tool to also leverage other components that are crucial for Cas9 activity,” says Dr. Laurence Wilson whose research focuses on computational genome engineering.
Specifically the off-target search is a compute intensive task traditionally reserved for researchers at large institutes with high-performance-compute infrastructure as every location in the 3 billion letter long genomic sequence needs to be investigated. GT-Scan2 democratizes the ability to find optimal sites by offering this complex computation as a cloud-service using AWS Lambda functions.
Scaling Instantaneously for Personalized Treatments
GT-Scan2 leverages the instantaneous scalability that the event-driven AWS Lambda service offers. This is crucial for personalized treatment, as complexity of the targeted gene can vary dramatically.
“The off-target search as well as the robustness analysis can be subdivided into independent, modular tasks that can run in parallel” says Aidan O’Brien who designed and implemented the system within weeks after its official Asia-Pacific launch in April this year at the AWS Summit 2016 attesting to the intuitive nature of the service. A typical job takes less than a minute and the variation between jobs range from 1 second to 5 minutes. This fast fluctuation in load over minutes rather than hours ruled out an EC2-based solution as new instances would come online too slowly to keep the runtime stable.
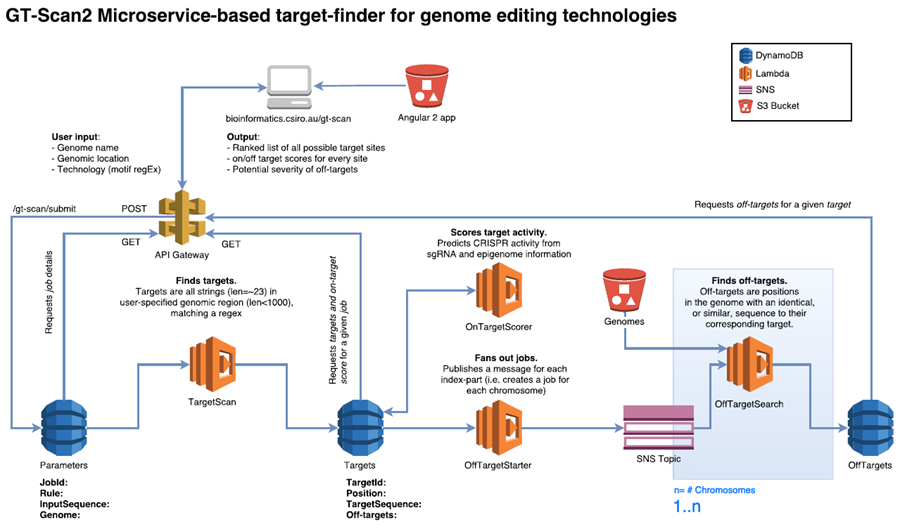
GT-Scan2 is served directly from S3 making it a static web app without server-side processing. It retrieves the dynamic content (such as job results and parameters) via API calls using API Gateway from a database (DynamoDB) using a JavaScript framework.
When a user submits a job, GT-Scan2 inserts the job parameters as an item into a DynamoDB table via an API call. This allows the solution to be freely scalable without creating a bottleneck. The database entry triggers the first Lambda function, which finds all putative CRISPR targets in the user-specified DNA sequence (fetched automatically upon user submission). Potential CRISPR target sites have fixed rules and can be easily found using a regular expression that completes in seconds and are inserted into a second DynamoDB table.
Adapting to leverage the power of Lambda-based microservices
All potential targets need to be evaluated for their off-target risk using the efficient string matching tool, Bowtie. Though Bowtie only requires a reduced representation of the 3 billion letter genomic sequence, the sizes of these index files exceed the storage limitation for each Lambda instance. “GT-Scan2 divides the genome into smaller blocks to fit the Lambda specifications” explains Adrian White (Research & Technical Computing, APAC) who supported the CSIRO team during development. For an average run, GT-Scan2 hence triggers 500-1000 individual Lambda functions, which simultaneously update the scores for the different putative targets in DynamoDB. During this process, the frontend is polling this table via API Gateway and updating the webpage as results come in, eliminating the need for server-side compute.
“AWS’s Lambda has given us a great framework to develop a future-ready software package able to support medical genome engineering applications,” says Dr. Bauer. “We are specifically impressed with the ability to instantaneously scale at run time by spawning more Lambda functions to cope with the varying complexity of the different genes.” Other benefits Dr. Bauer quotes include only paying for storage during periods of no use and jobs not competing with web server resources as the website is a static page with dynamic content updated through Angular 2 and the API Gateway, as well as not needing to maintain compute instances (security patches of OS).
“One of the best things about Lambda is that users will be able to easily swap-in different machine learning algorithms that are better suited for specific CRISPR applications” says Dr. Wilson.

The GT-Scan2 Team, from left, Denis Bauer, Laurence Wilson, Aidan O’Brien
“The computational genome engineering community is one of the early adopters of our AWS Lambda technology,” explains Dr. Mia Champion (Technical Business Development Manager, Scientific Computing). “GT-Scan2’s use of API Gateway and DynamoDB is a very neat solution to ensure scalability and their clever use of epigenomics really sets them apart from other recent applications using lambda to perform CRISPR searches. I am looking forward to seeing GT-Scan2 adopted in medical applications.”