AWS for Industries
At-scale Genomic Data Compression, Storage, and Access Using PetaGene on AWS: Reference Architecture
Analysis of large-scale genomic data is the foundation for developing new treatments that make personalized medicine a reality. However, access to these multi-petabyte datasets brings challenges in terms of managing storage and compute infrastructure. The scale and flexibility of AWS resources are ideal for such projects.
AWS has developed a Reference Architecture for compression and read back using PetaGene’s PetaSuite Cloud Edition and deployed it with a large biopharma client. By doing so, the client has reduced their storage footprint, sped up data ingress and egress, and accelerated analysis, while maintaining the data in a fully-accessible format. This blog describes this Reference Architecture and how to implement it.
Processing pipeline
PetaGene compression is run within the biopharma client’s genomic processing pipeline during the Compression and Ingestion step as shown in the figure below. The scope of this Reference Architecture only covers Compression and Ingestion with the other steps shown for context.
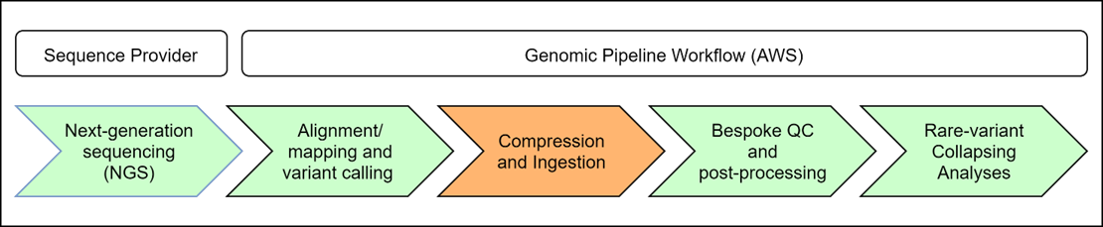
Figure 1: Processing Pipeline Example
Data from the Alignment/mapping and variant calling step arrives in variable size batches rather than as a constant stream. Once this step completes, output data is compressed using PetaGene and then ingested into a S3 bucket used for permanent storage as well a file catalog, which maintains metadata about each file. Once the Compression and Ingestion step is finished, the Bespoke QC and post-processing step runs a number of quality control checks and post-processing programs (peddy, verifybamid, STR expansion hunter, TelSeq, etc.), and finally, data is available to scientists for interpretation.
PetaGene’s compression software runs multi-instanced and multi-threaded and therefore scales to arbitrarily large batch jobs. There is no central server to create a bottleneck. PetaGene’s license manager only records billing and status information about the compressions, and presently can handle a compression request rate of more than 200,000 files per user per hour. PetaGene’s PetaSuite minimizes the cloud storage footprint and data transfer time, while maintaining maximum data access and processing performance.
The biopharma client initially analyzed 220,000 whole exomes sequences at a rate of 1,000 sequences per hour, resulting in 770 TB of BAM files. PetaGene’s PetaSuite lossless compression was then able to shrink their size by a factor of more than 4x at a rate of 10,000 files per hour, after which they were written to tiered long-term storage and subject to periodic retrieval. Using PetaGene’s PetaSuite, the client will reduce their storage and retrieval costs by more than $1 million over three years, even after taking into account the PetaGene fee, which is a fixed price per TB saved.
Subsequently, the client included the PetaGene compression step within the analysis pipeline, as described in detail below.
Architecture overview
The following diagram depicts an overall architecture that you can insert into a new or existing pipeline. Since PetaGene’s PetaSuite Cloud Edition runs well within a Docker container, we use AWS Batch and Elastic Container Services (Amazon ECS) to provision and auto-scale the compute and memory resources needed to perform the compression. We use AWS Simple Queue Service (Amazon SQS) to keep track of the work that needs to be performed, with a Lambda function that reads from the queue and then dispatches work to AWS Step Functions. Compression work can be instantiated in two ways: when the workflow that creates the file to be compressed finishes, the compression workflow is automatically started; or for compressions to be performed retroactively, a user can submit batches of compression jobs to run.
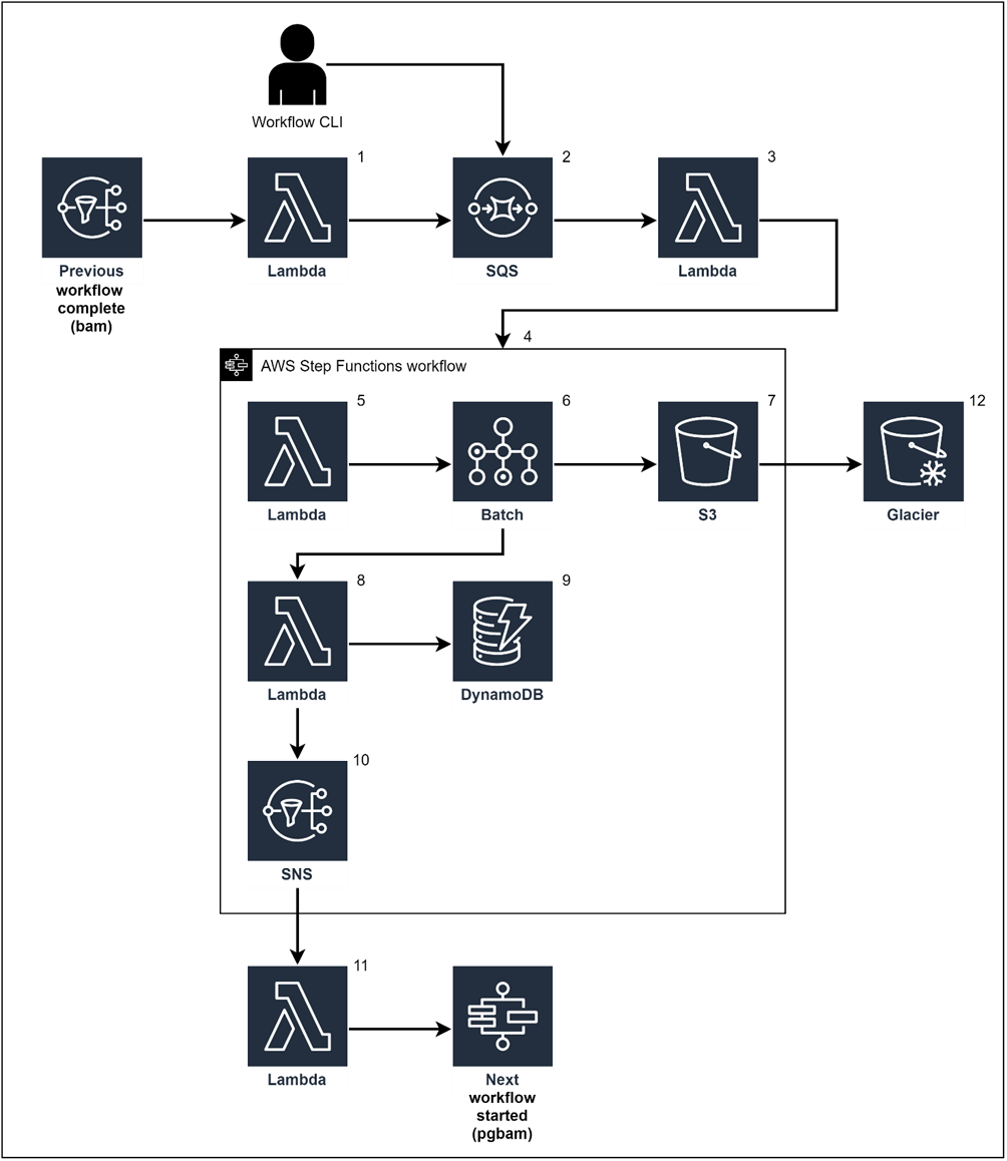
Figure 2 – AWS PetaSuite Architecture
Here is a step-by-step walkthrough of the architecture shown above, with a basic overview of how each AWS service fits together to perform compression:
- When the analysis workflow finishes, a message is written to a Simple Notification Service (SNS) topic. A Lambda function that subscribes to the topic writes a message to an Amazon SQS queue. For retro-active compression, a user can submit a list of files to be compressed via a simple CLI program with the CLI program sending one message to the queue for each file to be compressed.
- The SQS queue is a first-in-first-out (FIFO) queue, which processes the messages in order of occurrence.
- A Lambda function reads from the queue and invokes a Step Functions workflow, which coordinates the work to be performed for each compression.
- One Step Functions workflow is invoked for each file to be compressed.
- The first step of the compression workflow is a Lambda function, which writes the information needed by the batch job to a temporary JSON file in Amazon S3. This file contains all of the information that will be needed by PetaGene’s PetaSuite to perform the actual compression, such as the name of the file and the location of the file in S3.
- AWS Batch is then invoked. The first step of the batch job running in a Docker container, which has PetaGene’s PetaSuite installed, is to retrieve the JSON file created in the step above. The batch job then runs a wrapper script, which calls PetaSuite with the parameters needed to run.
- The last step of the batch job is to upload the compressed files to S3.
- Now that we have the compressed files stored in S3, a Lambda function updates a file catalog, which records information about each file.
- The file catalog is held in a DynamoDB table and stores one item for each file including attributes for file name, file size, file type, and the location of the file in S3. Since PetaGene’s PetaSuite generates a checksum for each file that it creates, we store the checksum in the file catalog as well.
- The last step of the workflow writes a message to an SNS topic.
- A Lambda function subscribes to the topic and writes a message to another SQS queue to be used to invoke the next workflow in the pipeline.
- After all downstream processing is complete, tags are applied to the S3 object, which enable lifecycle management rules that move the compressed files to Amazon S3 Glacier for long-term storage.
PetaGene’s PetaSuite initially performed retro-active compression of data that had already been processed. Now, the compression element is triggered automatically in the production pipeline handling new data, using the architecture described above.
Getting started
The architecture shown above can be deployed using AWS CloudFormation. We also create a container with PetaGene’s PetaSuite installed, and store this container in Elastic Container Repository (Amazon ECR). The numbers in parentheses below refer to the processing steps described above.
Container
Build a Docker container and upload to Amazon ECR. PetaGene’s PetaSuite is added with a few lines in the Dockerfile. Assuming Ubuntu as the base image:
COPY --from=builder /source_files/petasuite-cloud-edition-proxy_amd64.deb /tmp/
Lambda functions
All of our Lambda functions are written in Python and we use the boto3 library to make calls to AWS services.
Function (1) subscribes to an SNS topic, which indicates that the previous workflow has been completed. In our case, the previous workflow is the analysis process that generates a BAM file. The Lambda function reads the SNS messages and writes a new message to a SQS queue (2) with the message body containing the details of the file to be compressed.
Function (3) is triggered by a message arriving in the SQS queue (2) and invokes the compression workflow StepFunction. The input to the StepFunction is the message body content.
Function (5) writes the message content to a JSON file, which is stored in a known location in S3. This provides the batch job with a convenient method for receiving its work instructions.
Function (8) writes file metadata to a file catalog once the compression batch job is finished. The input to this function is the various metadata attributes that we want to store for each file. In our case, this includes file name, size, type, checksum, data created, and S3 bucket and key for the file. When finished, this function writes a message to an SNS topic (10) to indicate that the compression workflow has been completed.
Function (11) is an optional function, which can be used to trigger the next workflow in the pipeline. If you don’t have any further workflows to run, this function is not needed.
Batch script
When the batch container starts, the entry point is a Python script that retrieves the work instructions written to an S3 object by Function (5). This script includes a function, which uses a Python subprocess to invoke PetaGene’s PetaSuite:
The main function of this script does the following:
AWS CloudFormation
We use AWS CloudFormation to deploy all of the required infrastructure. This includes the following:
- File store S3 bucket with associated Lifecycle policies for storing compressed files and other files produced by the pipeline.
- SQS queue for incoming work management.
- Lambda functions as described above.
- SNS topic for notification of when the compression workflow completes.
- Batch components including: compute environment, queue, and job definition.
- DynamoDB table used to store file level metadata.
Results
Table 1: Whole Exome and Whole Genome data compression achieved
| Whole Exomes | Whole Genomes | |
| Files processed | 218,761 | 2,988 |
| Input file type | BAM | |
| Input file size | 3.5 GB | 70 GB |
| Compression reduction | 4.2x | 4.2x |
| Output file size | 0.8 GB | 16.7 GB |
| Output file type | Compressed BAM in accessible format | |
| Average time to compress and validate one file | 5 mins | 80 mins |
| EC2 instance type used | m5.2xlarge (8 vCPUs, 32 GB mem) |
m5.2xlarge (8 vCPUs, 32 GB mem) |
| Configured rate per hour | 10,000 | 625 |
The “Configured rate per hour” is adjustable and is dependent on (i) the size of the EC2 instance since PetaGene’s PetaSuite compression is multi-threaded, and (ii) the number of EC2 instances that you have running concurrently. The architecture imposes no limit on this rate and can be configured to achieve your desired objective. For the client project described in this blog, the goal was to be able to compress all of the files in under two days. This goal was achieved using m5.2xlarge instances and running approximately 850 instances concurrently.
Conclusion
Specific features of PetaGene’s PetaSuite compression mean that the benefits of compression can be realized throughout the analysis process.
- Thanks to random streaming access, there is no need to transfer whole files from S3 object storage to EC2 instances before starting compute operations. This reduces data analysis time by eliminating the need to download and decompress files before use.
- The serverless architecture using AWS Step Functions and AWS Batch quickly scales compute instances up and down according to demand to minimize compute costs.
- PetaGene’s unique compression algorithms are optimized for genomic data. The average compression saving is 76% with a tight distribution, better than a 4x file size reduction.
- PetaGene’s transparent access means that compressed data can be deeply embedded in the analysis process without having to re-engineer standard cloud or bioinformatics technologies to accommodate compressed data.
Collaboration is an important element of advancing genomics research. Thanks to PetaGene’s freely available access library PetaLink, the compressed data can be shared with third parties, who can read them as regular BAM files without needing to decompress first. This AWS-based Reference Architecture allows you to build partnerships and collaborate with other genomics innovators.