AWS for Industries
Global OHDSI COVID-19 Study-A-Thon Produces Important Insights
What happens when a pandemic cancels your annual scientific meeting? You improvise.
The Observational Health Data Sciences and Informatics (OHDSI, pronounced “odyssey”) community planned to celebrate recent research initiatives and discuss future efforts during the annual European Symposium at Oxford University in late March. COVID-19 had other plans. Opting to flatten the curve, symposium organizers cancelled the in-person meeting, but they didn’t stop there.
Instead, the OHDSI community launched a COVID-19 global, virtual study-a-thon March 26-29, believing that a network of people who valued both collaboration and open science could make a meaningful impact on the current global pandemic. As a community, OHDSI is an open science initiative of individuals who volunteer their time and talents for the shared goal of improving healthcare through observational research.
With COVID-19 spreading rapidly and exhausting health systems around the world, OHDSI knew it had the unique opportunity to respond to this challenge. The community designed and began executing studies on an international set of observational health databases (including insurance claims and electronic health records) to aid decision-making during the current COVID-19 pandemic. More than 330 people from 30 nations registered to collaborate in this 88-hour virtual study-a-thon, which concluded March 29 with a global presentation from multiple study leads to announce both designs and preliminary findings. Results are currently being evaluated and papers are actively being submitted to journals for peer review.
One study is the first large-scale characterization of COVID-19 patients hospitalized in both the United States and Asia involving six databases with COVID-19 patients located in both the U.S. and South Korea. Another study is the largest ever conducted on the safety of hydroxychloroquine and was designed and executed across an international set of health databases. This study of more than 950,000 patients from the USA, England, Germany and South Korea focuses on the overall safety profile of hydroxychloroquine, a drug currently being evaluated as a potential treatment for COVID-19. A pre-print of this analysis is available on medRxiv, and it highlights the risks involved with combination therapy of hydroxychloroquine and azithromycin. A third study designed the first prediction model externally validated on COVID-19 patients to support triage decisions in an effort to ‘flatten the curve’. This model determines whether a patient is likely to require hospitalization based on their symptoms and was developed against US data and then tested on South Korean data.
“It was a tough blow initially to not host the OHDSI European Symposium at Oxford, but I was so proud and honored to help lead the virtual study-a-thon. It was thrilling to bring together a global community in response to this pandemic, and it has been inspiring to continue collaborating on COVID-related work that will make a real difference. Studies on the safety profile of hydroxychloroquine and a multi-institution characterization of hospitalized COVID patients are early examples of findings that we are excited to share with the healthcare community.”
Dani Prieto-Alhambra, MD, MSc(Oxf), PhD, Professor of Pharmaco- and Device Epidemiology University of Oxford
An International Health Data Network
All of these studies were conducted across an international network of 37 health databases without ever sharing patient-level data. The OHDSI community has been working toward this goal by producing open data standards and open-source analytics tools for more than 10 years. Using the OHDSI tools, you can build cohorts of patients, analyze incidence rates for various conditions, and estimate the effect of treatments on patients with certain conditions. You can also model health outcome predictions using machine learning algorithms.
“OHDSI has always championed the principles of open science through supporting open-community data standards, developing open-source analytics tools, and coordinating open-research studies. The COVID-19 pandemic has demonstrated why these principles matter so much. Through open collaboration, our community is working to deliver answers to some of the most pressing questions shared by governments and healthcare leaders. While Columbia University is proud to serve as the central coordinating center of OHDSI, it is our global community of collaborators, including Amazon Web Services, who are showing both the power and potential of open science.”
George Hripcsak MD, MS, leader of the OHDSI Coordinating Center, Vivian Beaumont Allen Professor and Chair of Columbia University’s Department of Biomedical Informatics and Director of Medical Informatics Services for NewYork-Presbyterian Hospital/Columbia Campus
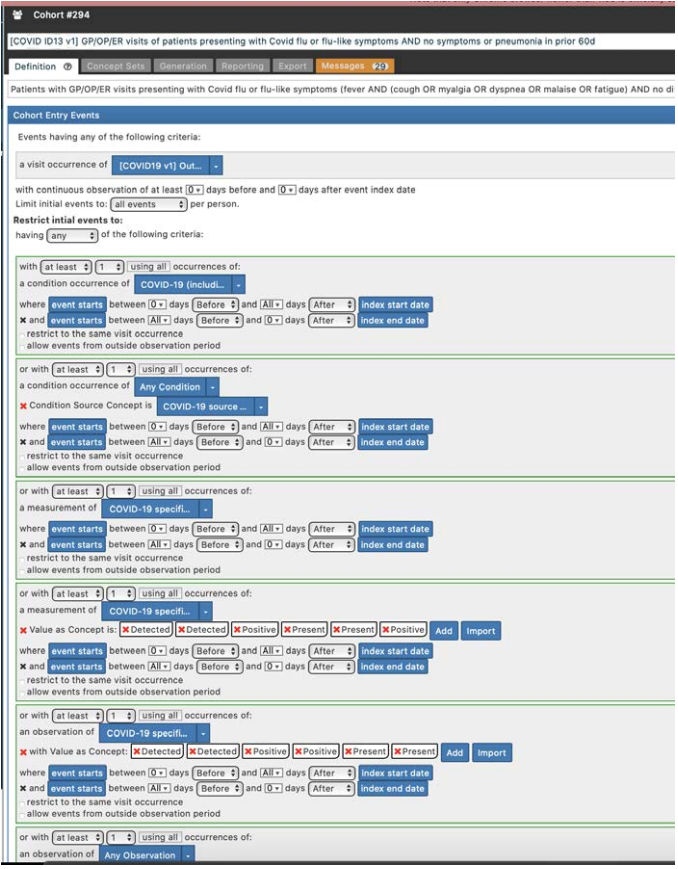
One of the OHDSI tools, Atlas, is a web application where researchers can define the phenotype criteria for patients to be considered as a part of a cohort. To do this, they use standard vocabulary codes that can be applied to any OHDSI system, regardless of where the source data came from. Above is an image showing part of the definition used to define patients with COVID-19 or flu symptoms who had no symptoms 60 days prior.
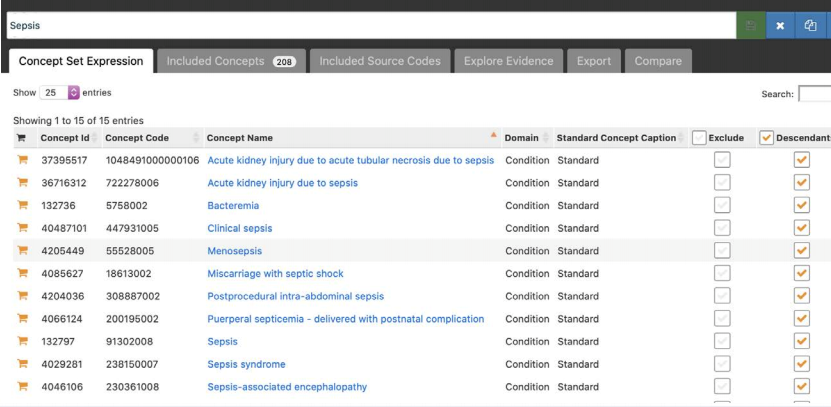
OHDSI also provides a standardized vocabulary that allows data encoded in over 100 different healthcare ontologies (like SNOMED CT, ICD-9, ICD-10, RxNorm, READ, and many more) to be analyzed, easing the ability to execute global health studies. This allows researchers to define generalized and portable definitions for conditions, procedures, and cohorts of patients that can be used by other organizations without needing to see that organization’s data.
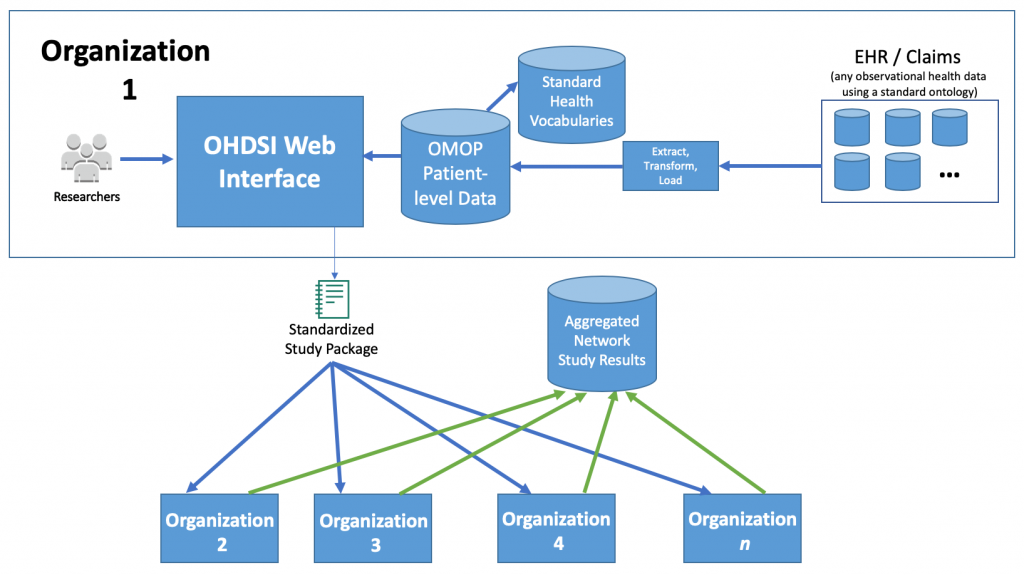
Because OHDSI tools are built to be compatible with data in a standardized data model, called the OMOP Common Data Model, you can create sharable analytics packages. This enables anyone with an OMOP formatted data set to run the analysis and share back the results data without ever exposing their patient-level data. This is how the OHDSI community was able to quickly craft and execute analysis packages for so many important health questions against such a large number of data sets in under 4 days. The OHDSI community collaborated in a publicly accessible instance of Atlas that contained no real patient data to define hundreds of phenotype definitions and analysis packages that would be distributed to the various participating institutions. These institutions then received these standard analysis packages, executed them against their patient data in the OMOP Common Data Model, and returned the results. OHDSI tools also have data quality checks that help participants to ensure the accuracy of their results.
The public environment used for collaboration was deployed in just a few hours using the OHDSIonAWS project. This project allows organizations to quickly get started with the OHDSI toolset and OMOP Common Data Model by automatically deploying a best-practices OHDSI environment. Some of the participating organizations also ran their data analysis using AWS data analytics services, like Amazon Redshift.
“It was really exciting to see the entire OHDSI community rally around a common cause: to collaboratively generate reliable evidence that could meaningfully inform health decisions around COVID-19. The OHDSI COVID study-a-thon demonstrates what’s possible when you bring together smart minds, scientific best practices, and a network of data from around the world. This collaboration wouldn’t have been possible without the support of AWS for providing computational resources and hosting the OHDSI analytics infrastructure to enable researchers to work together on a common platform. I am eager to continue this important partnership and excited about where the future journey together will take us.”
Patrick Ryan, PhD, Vice President, Observational Health Data Analytics at Janssen Research and Development, Assistant Professor, Adjunct; Department of Biomedical Informatics Columbia University Medical Center
You can join the journey
The OHDSI tools and data standards are free and completely open source. You can take advantage of them privately to study your own patient data and gain important insights to help improve clinical treatment. You can also, optionally, join the OHDSI Data Network and decide to participate in global health studies like these ongoing OHDSI COVID-19 research investigations.
“The European Health Data and Evidence Network project (EHDEN) is bringing together data across Europe, but it’s clear that we need to come together across Europe, North America, Asia and all parts of the world to generate reliable evidence that can be used to fight this pandemic. Our work in the EHDEN Academy, which has received critical support by AWS, will enable a global community to ‘join the journey’ and expand the scope of OHDSI’s impact on health and healthcare, especially in response to our current global crisis.”
Peter Rijnbeek, PhD, associate professor health data science at the department of medical informatics of the Erasmus University Medical Center, Rotterdam, The Netherlands. He is the coordinator of the European Health Data and Evidence Network (EHDEN) project.
You can get started by
- Become familiar with OMOP CDM and OHDSI methods via the Book of OHDSI (book.ohdsi.org).
- Get hands-on training via the newly launched EHDEN Academy (https://academy.ehden.eu/course/index.php), which provides short online courses on OHDSI in a Box (OHDSI on AWS), OMOP CDM and Standardized Vocabularies, Process for Extract, Transform and Load and Infrastructure Set-up.
- Get involved in the conversation via the OHDSI Forums (forums.ohdsi.org).
- Visit the OHDSI Studies Repo (http://www.github.com/ohdsi-studies/) to see the code base for active research investigations including the COVID-19 work and other community research initiatives.
- Reach out to contact@ohdsi.org if you have specific questions. The OHDSI community will help direct you to the collaborators who lead the workgroups you’re interested in.