AWS Big Data Blog
Analyzing healthcare FHIR data with Amazon Redshift PartiQL
Healthcare organizations across the globe strive to provide the best possible patient care at the lowest cost. In a patient’s care journey, multiple organizations are often involved, including the healthcare provider, diagnostic labs, pharmacies, and health insurance payors. Each of these organizations needs to exchange health data efficiently with the others to ensure care continuity and reimbursement.
The healthcare industry has adopted data exchange standards, many of which are defined by Health Level Seven International (HL7), for several decades. HL7v2, a pipe-delimited data format developed three decades ago, is still in use today despite not conforming to modern best practices for communicating between systems, such as with RESTful APIs. Naturally, these shortcomings can complicate interoperability. More recently, HL7 introduced FHIR (Fast Healthcare Interoperability Resources) to help solve some of the complexity and pave the way for healthcare organizations to modernize how they exchange information. FHIR is built around resources that logically organize healthcare information in a structured but fully extensible format. FHIR is quickly becoming the standard for information exchange in the healthcare industry; for example, the United States’ Centers for Medicare & Medicaid Services (CMS) recently announced the Interoperability and Patient Access final rule (CMS-9115-F), which adopts FHIR as the standard for exchanging health data.
In addition to exchanging information with other entities, healthcare organizations are recognizing the intrinsic value of their own health data flowing within their systems. By bringing analytics to their own health and operational data, leading healthcare organizations are now improving care quality, patient experience, and cost. However, existing tooling for data visualization, statistical analysis, and machine learning often relies on relational schemas that can be easily transformed into vectorized inputs. The majority of existing analytics infrastructure relies on this “flat” storage and presentation of data assets; this can be challenging given FHIR’s heavily nested JSON structure. In addition, data scientists need to consume FHIR format from multiple sources and connect them with each other and existing relational data that resides in existing databases.
In this post, we walk through how to use JSON Schema Induction with Amazon Redshift PartiQL to simplify how you analyze your FHIR data in its native JSON format. Although this post focuses on a simple analysis of claims data, this approach can help data scientists and data analysts reduce the manual work and long cycles of data processing when analyzing patient data by querying and running statistical analysis that is required day to day.
JSON Schema Induction and PartiQL
There are multiple ways to organize and query healthcare data on AWS. One such way is to flatten and normalize the nested JSON FHIR documents so that it’s usable in traditional relational schema. However, such an exercise delivers a subpar final model that results in hundreds of tables with thousands of columns that aren’t naturally extensible the same way FHIR is designed. In addition, consumption of the data from such a relational model is time-consuming and expensive. Alternatively, you can leave the FHIR resources in their hierarchical form and eliminate the need to invest and support a complex ETL pipeline. This approach, however, presents separate challenges because the existing analytics tools data scientists are most comfortable and productive with aren’t often well suited to interrogate deeply hierarchical data structures.
In August, 2019, AWS announced PartiQL, an open source SQL-compatible query language that makes it easy to efficiently query data regardless of where or in what format it is stored. This technology simplifies how data scientists can use SQL to directly query FHIR resources. Our approach is different from using nested types to simplify and optimize query processing. We focus on native hierarchical schema that tends to be much more complex, with many nesting levels and optional structures.
Although Amazon Redshift PartiQL is an enabling technology to query and explore, analysts and scientists also require an understanding of the underlying structure they are interacting with. The FHIR standard incorporates descriptions of data elements as first-class members and presentation of this context alongside the data itself promotes a richer understanding. Despite existing tools having the ability to infer a schema from data (such as Apache Spark and AWS Glue crawlers), they can’t incorporate additional structural information about the data, as found in the FHIR standard. These tools also don’t offer immediate DDL generation, which is very useful to create schema files that define your catalog structure.
As the schema becomes more complex, it’s harder to devise the DDL for it manually, and using our schema induction library becomes necessary. Programmatically inferring the JSON schema is a mechanism that you can use to query FHIR, or any set of JSON documents, in their native structure. We present the tooling required for JSON Schema Induction and provide step-by-step examples to help you get started querying FHIR resources. In this post, we focus on healthcare data in the FHIR format, but our approach for analyzing hierarchical data in JSON or XML formats is applicable to many other data standards in other domains.
To address these challenges and overcome the shortcomings of existing approaches, we have introduced a new open-source library for inferring a schema from a set of JSON documents. As a result of this inference, we can to generate table definitions that significantly streamline the ability to efficiently process FHIR data directly. This library combines the induced schema from the sample data with the descriptive information from the FHIR standard to generate a rich DDL for Amazon Redshift and a JSON tree for UI visualization.
Solution overview
The following diagram illustrates the architecture of the solution.
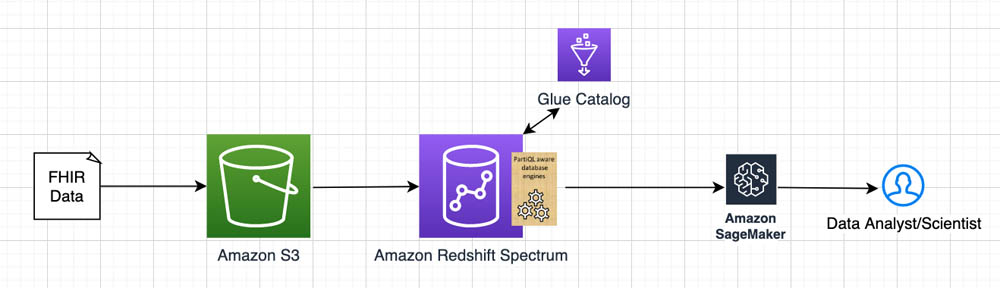
In this post, we demonstrate how to use our open-source library with conjunction of Amazon Redshift PartiQL queries and Amazon Redshift Spectrum, which enables you to directly query and join data across your data warehouse and data lake. Our approach allows you to design a semi-relational schema, where scientists can quickly combine relational and non-relational data from multiple resource tables.
We use PartiQL in Amazon Redshift Spectrum over data stored in Amazon Simple Storage Service (Amazon S3). Amazon S3 is an object storage built to store and retrieve any amount of data, structured or unstructured, while providing extreme durability and availability, and infinitely scalable data storage infrastructure at very low costs. Amazon Redshift Spectrum runs complex SQL queries directly over Amazon S3 storage without loading or other data preparation, and AWS Glue serves as the meta-store catalog for the Amazon S3 data. That allows us to run PartiQL queries on Amazon S3 prefixes containing FHIR resources stored as JSON or Parquet files. We use Jupyter notebooks to illustrate how to build and use the Schema Induction tool, and for test data we use the public dataset from Fhirbase.
Querying FHIR using PartiQL
To provide a short introduction to PartiQL and illustrate the power of PartiQL, we show a few examples of PartiQL queries over FHIR data. Each example shows different aspects of query functionality varying from simple to aggregate queries. Although you can use this approach for myriad FHIR resources, we focus on claims data as an illustrative example. If you’re already familiar with PartiQL language, you can skip this section.
Assume that we have millions of claim documents as illustrated in claim-example-cms1500-medical.json and in the following code. We want to process attributes regarding ID (line 3), status (line 14), patient (lines 24–26), and diagnosis (lines 58–69):
Creating the claims table DDL
To run queries with Amazon Redshift Spectrum, we first need to create the external table for the claims data. The claims table DDL must use special types such as Struct or Array with a nested structure to fit the structure of the JSON documents. For the FHIR claims document, we use the following DDL to describe the documents:
Because each column might be a highly nested JSON structure, this structure requires many levels of type definitions and optional attributes, as shown in lines 9–14. The definition of the preceding table works for a certain type of document in our sample. A table definition that can satisfy any claim document as defined by the FHIR JSON schema is even more complex. This is because the FHIR schema aims to capture any possible medical information, not just the existing data you have, and therefore is more complex than what you currently need.
Traditionally, creating a DDL table is a one-time operation performed by the database administrator, but devising DDL manually isn’t intuitive when it comes to a complex hierarchical dataset. Therefore, we created a new tool: the Schema Induction Tool, which is an open-source library that infers a schema out of a collection of documents and produces rich structure information of the collection of documents scanned.
Generating DDL with the Schema Induction Tool
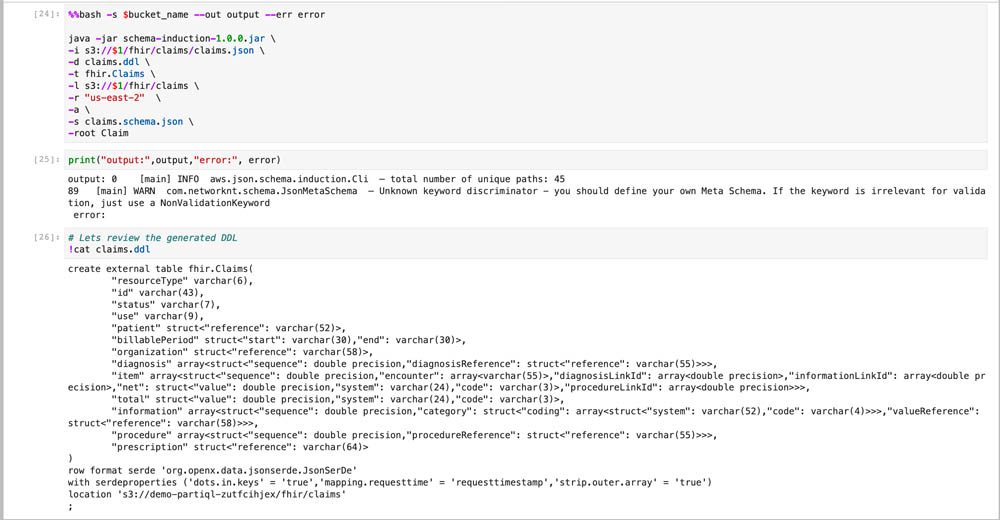
The Schema Induction Tool is a java utility that reads a collection of JSON documents as stream, learns their common schema, and generates a create table statement for Amazon Redshift Spectrum. In addition, if the documents adhere to a JSON standard schema, the schema file can be provided for additional metadata annotations such as attributes descriptions, concrete datatypes, enumerations, and more. The JSON standard provides a comprehensive data structure for all possible variations of the data, whereas the enriched inferred schema only considers the structure that it has seen from the documents, and doesn’t explode the DDL statement with all the theoretical possibilities that the JSON schema defines. It is therefore imperative to provide a rich and heterogeneous set of documents that can provide the total possible paths in the current data. Because the utility uses stream processing, you can run it on a large volume of documents if time permits.
You can build the code yourself from the GitHub repo, or download a release artifact from Maven central.
The CLI syntax of the command is as follows:
The utility generates a DDL output file that contains the Create Table statement as defined by the ddl switch, and optionally a file that contains the hierarchical schema of the document collection as defined by the schema switch. Finally, a stat file is generated that contains the total number of occurrences of each element from the input documents collection.
Building the Induction Tool
We can now put it all together so we can analyze FHIR Claims data. We assume you have access to AWS account and also to a Linux terminal where you can access the data on Amazon S3 and run commands. We use Amazon SageMaker to host our Jupyter notebook.
- Build the Schema induction tool with the following code:
You use Amazon SageMaker to generate the Create Table statement for the claims.
- Create an Amazon SageMaker notebook on the Amazon SageMaker console.
- Select the notebook and choose Open JupyterLab.

- Copy the example data from your code folder
schema-induction/target/schema-induction-1.0.0.jarto the notebook by dragging the file and dropping it into the left panel.

- Copy the example data from your code folder
data/claims.jsonto the notebook. - Copy the notebook document from your code folder
notebooks/fhir-Partiql-notebook.ipynb. - Open the notebook, set up the variables in the first cell per your account, and run the first cells to download and build the schema induction tool.

Preparing the data
Now the induction tool is ready and we can perform the schema induction.
- Run the next cells to download and check the test data.

- Check that the schema induction tool is ready and review the help guide.

Creating the FHIR table on Amazon Redshift
- Run the tool and review the induced DDL.
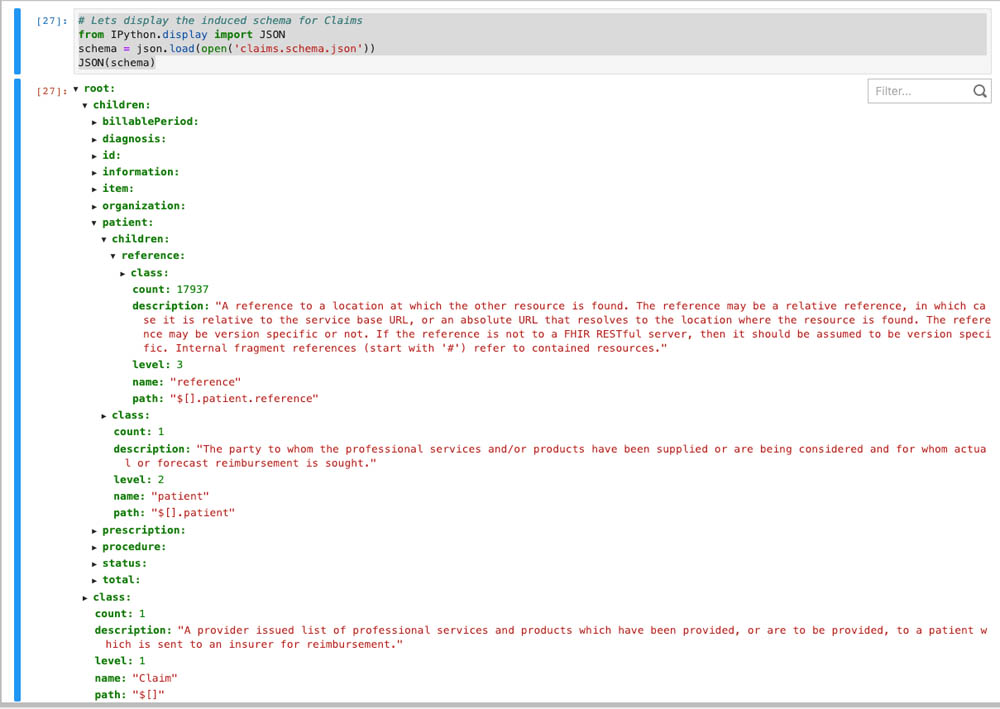
You can explore the data structure using the tree control display by running the following cell:
- On the AWS Glue console, create a database called
fhirand leave location empty. - Create an AWS Identity and Access Management (IAM) role that allows your Amazon Redshift cluster to access the S3 bucket and name it
fhir-role.
For more information about creating an Amazon Redshift Spectrum role that can access your S3 buckets, see Authorizing Amazon Redshift to access other AWS services on your behalf.
- On your Amazon Redshift cluster, make sure you added the IAM role so you can run the queries and access Amazon S3 and AWS Glue and that the status shows as
in-sync.

You can now run your PartiQL queries.
- Open the SQL Editor and connect to your database (in this use case, we use the Amazon Redshift Query Editor).
- Create the external schema in Amazon Redshift by entering the following code:
For more information about creating external schema in Amazon Redshift, see CREATE EXTERNAL SCHEMA.

- Create the external table by copying the
Create TableDDL from your notebook into your query and running it.

- Run your first PartiQL query on a new Query
Querying claims data
Claims are used by providers, payors, and insurers to exchange financial information and supporting clinical information regarding the provision of healthcare services with payors for reporting to regulatory bodies and firms that provide data analytics. In this section, we provide queries as examples of the analyses you can run.
The following query scans all documents in the users.Claims table and retrieves information from each claim.
The claim.diagnosis is an array that might contain multiple objects; it is therefore referenced in the SQL From clause and joined with the parent document implicitly, then attributes from the diagnosis element can be retrieved in the SELECT clause. This native SQL approach to un-nest arrays is one of the cornerstones of the PartiQL language. The query also uses the dot notation to access attributes in nested structures, such as c.patient.reference, which accesses the reference attribute inside the patient structure that is in the claim document.
Assuming there are claim documents without diagnosis information, and we want to retrieve the claims even if they have no diagnosis, we want a more permissive join by turning it into a left join. See the following code:
You don’t need to specify the join keys because the join key is essentially the parent-child relationship of the claim and diagnosis structures.
The following query performs a simple aggregation over the claims data:
It uses the c.patient.reference as the grouping key and the c.total.value as the aggregated value.
The following code queries all the patients’ claims:
The following screenshot shows the query output.
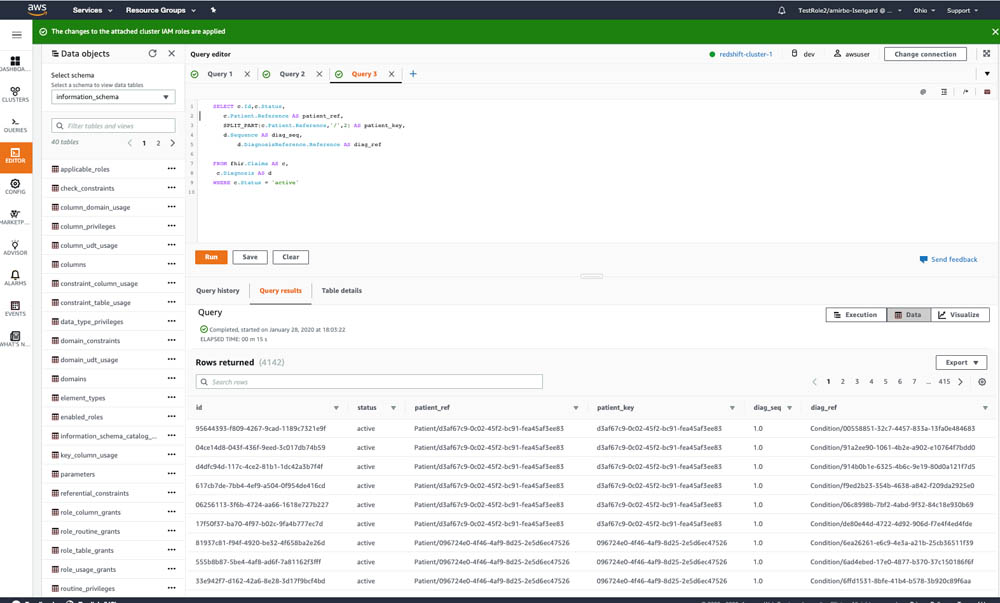
Congratulations, you have successfully queried FHIR data in Amazon Redshift Spectrum using PartiQL language.
Conclusion
The FHIR standard looks promising to do great things for healthcare and although it has limitations, we believe the benefits are here to stay. You can directly analyze FHIR format without creating massive normalized design, and can leapfrog over competitors to save tremendous investments in data modeling and ETL maintenance. This post shows you the power of Amazon Redshift PartiQL and how your data science and data engineer team can handle complex data formats like FHIR easily with automation. Adopting these technologies can bring new innovation to the healthcare world and unlock new benefits and cost-optimization.
If you have any questions, please leave your thoughts in the comments section. We look forward to your feedback on our new open-source Schema Induction Tool in GitHub.
About the Authors
 Amir Bar Or is a Principal Data Architect at AWS Professional Services. After 20 years leading software organizations and developing data analytics platforms and products, he is now sharing his experience with large enterprise customers and helping them scale their data analytics in the cloud.
Amir Bar Or is a Principal Data Architect at AWS Professional Services. After 20 years leading software organizations and developing data analytics platforms and products, he is now sharing his experience with large enterprise customers and helping them scale their data analytics in the cloud.
 Dr. Aaron Friedman a Principal Solutions Architect at Amazon Web Services working with healthcare and life sciences startups to accelerate science and improve patient care. His passion is working at the intersection of science, big data, and software. Outside of work, he’s with his family outdoors or learning a new thing to cook.
Dr. Aaron Friedman a Principal Solutions Architect at Amazon Web Services working with healthcare and life sciences startups to accelerate science and improve patient care. His passion is working at the intersection of science, big data, and software. Outside of work, he’s with his family outdoors or learning a new thing to cook.