AWS HPC Blog
Category: Industries
How Evolvere Biosciences performs macromolecule design on AWS
In this blog post, we catch a glimpse into drug discovery to see how Evolvere Biosciences has deployed a customized architecture w/ AWS Batch and Nextflow to quickly and easily run its macromolecule design pipeline.
BioContainers are now available in Amazon ECR Public Gallery
Today we are excited to announce that all 9000+ applications provided by the BioContainers community are available within ECR Public Gallery! You don’t need an AWS account to access these images, but having one allows many more pulls to the internet, and unmetered usage within AWS. If you perform any sort of bioinformatics analysis on AWS, you should check it out!
Optimize Protein Folding Costs with OpenFold on AWS Batch
In this post, we describe how to orchestrate protein folding jobs on AWS Batch. We also compare the performance of OpenFold and AlphaFold on a set of public targets. Finally, we will discuss how to optimize your protein folding costs.
A serverless architecture for high performance financial modelling
Understanding deal and portfolio risk and capital requirements is a computationally expensive process that requires the execution of multiple financial forecasting models every day and in often in real time. This post describes how it works at RenaissanceRe, one of the world’s leading reinsurance companies.
Benchmarking NVIDIA Clara Parabricks Somatic Variant Calling Pipeline on AWS
Somatic variants are genetic alterations which are not inherited but acquired during one’s lifespan, for example those that are present in cancer tumors. In this post, we will demonstrate how to perform somatic variant calling from matched tumor and normal genome sequence data, as well as tumor-only whole genome and whole exome datasets using an NVIDIA GPU-accelerated Parabricks pipeline, and compare the results with baseline CPU-based workflows.
Data Science workflows at insitro: how redun uses the advanced service features from AWS Batch and AWS Glue
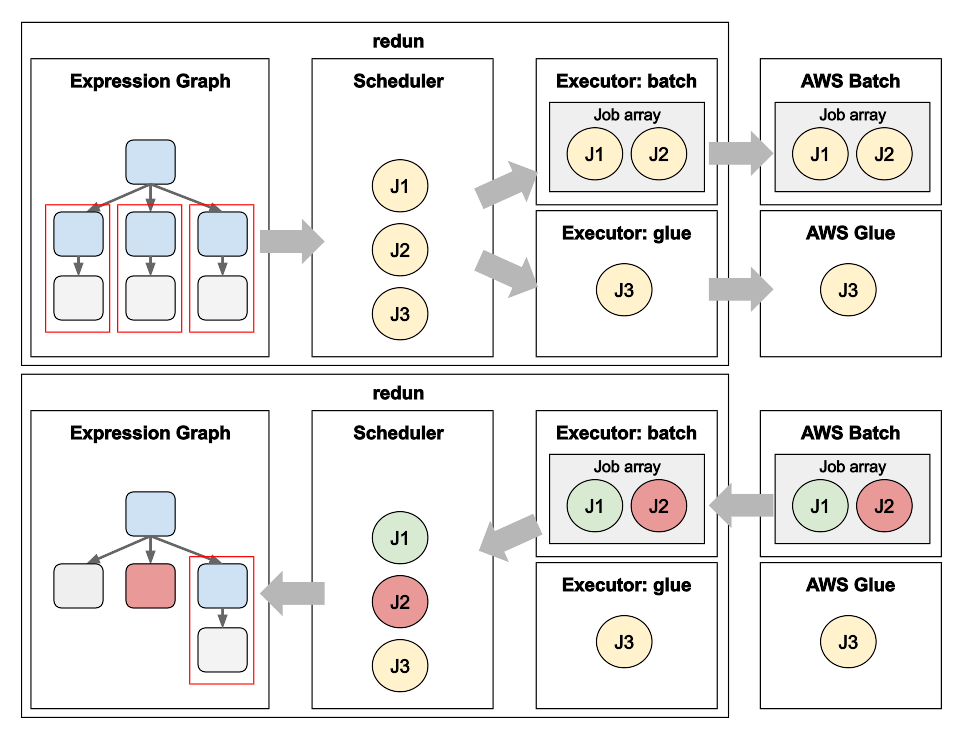
Matt Rasmussen, VP of Software Engineering at insitro, expands on his first post on redun, insitro’s data science tool for bioinformatics, to describe how redun makes use of advanced AWS features. Specifically, Matt describes how AWS Batch’s Array Jobs is used to support workflows with large fan-out, and how AWS Glue’s DynamicFrame is used to run computationally heterogenous workflows with different back-end needs such as Spark, all in the same workflow definition.
Data Science workflows at insitro: using redun on AWS Batch
Matt Rasmussen, VP of Software Engineering at insitro describes their recently released, open-source data science framework, redun, which allows data scientists to define complex scientific workflows that scale from their laptop to large-scale distributed runs on serverless platforms like AWS Batch and AWS Glue. I this post, Matt shows how redun lends itself to Bioinformatics workflows which typically involve wrapping Unix-based programs that require file staging to and from object storage. In the next blog post, Matt describes how redun scales to large and heterogenous workflows by leveraging AWS Batch features such as Array Jobs and AWS Glue features such as Glue DynamicFrame.
Creating a digital map of COVID-19 virus for discovery of new treatment compounds
Quantum physics and high-performance computing have slashed research times for a consortium of researchers led by Qubit Pharmaceuticals. This post describes the discovery of chemical substances that may lead to new COVID-19 treatments in only six months using cloud technology.
How we enabled uncompressed live video with CDI over EFA
We’re going to take you into the world of broadcast video, and explain how it led to us announcing today the general availability of EFA on smaller instance sizes. For a range of applications, this is going to save customers a lot of money because they no longer need to use the biggest instances in each instance family to get HPC-style network performance. But the story of how we got there involves our Elastic Fabric Adapter (EFA), some difficult problems presented to us by customers in the entertainment industry, and an invention called the Cloud Digital Interface (CDI). And it started not very far from Hollywood.
Running a 3.2M vCPU HPC Workload on AWS with YellowDog
OMass Therapeutics, a biotechnology company identifying medicines against highly validated target ecosystems, used Yellowdog on AWS to analyze and screen 337 million compounds in 7 hours, a task which would have taken two months using an on-premises HPC cluster. YellowDog, based in Bristol in the UK, ran the drug discovery application on an extremely large, multi-region cluster in AWS with the AWS ‘pay-as-you-go’ pricing model. It provided a central, unified interface to monitor and manage AWS Region selection, compute provisioning, job allocation and execution. The entire workload completed in 65 minutes, enabling scientists to start work on analysis the same day, significantly accelerating the drug discovery process. In this post, we’ll discuss the AWS and YellowDog services we deployed, and the mechanisms used to scale to 3.2m vCPUs using multiple EC2 instance types across multiple regions in 33 minutes, running at a 95% utilization rate.