AWS for Industries
Building a GPU-enabled CryoEM workflow on AWS
Cryo-electron microscopy (CryoEM) of non-crystalline single particles is a biophysical technique that can be used to determine the structure of biological macromolecules and assemblies. The technique is rapidly becoming the go-to technique in the life-sciences drug discovery process for understanding protein structure. Processing data/images coming from a CryoEM microscope demands a scalable storage for large datasets (median size per sample 1.2 TB) and a CPU/GPU enabled hybrid environment for performing computations. The complexity of the analysis provides an ideal use case for AWS Cloud services.
In this blog post, we discuss how AWS services provide an agile, modular, and scalable architecture to optimize the CryoEM workflows thus providing a pathway to wider adoption of CryoEM as a standard tool for structural biology
The Challenge
The 2017 Nobel Prize in Chemistry was shared by three distinguished scientists for a microscope technology (Cryo-electron microscopy or CryoEM) that is revolutionizing scientific discovery. CryoEM provides the ability to build detailed 3D models of biomolecules at near-atomic scale. It works using a 3-dimensional microscope imaging method that uses a transmission electron microscope (TEM) to visualize biological samples in a frozen hydrated near-native state. A subspecialty technique called Single-Particle CryoEM is being used for a wide range of biological samples – taking thousands and sometimes millions of images of molecules embedded in a thin film of ice with the TEM, then using computational approaches to classify and average these images into a 3D volume.
To fully deliver on the promise of CryoEM technology, researchers need timely access to both data and computational power. They need an IT infrastructure that is capable of handling the data growth, as well as enormous demand on computing using highly specialized packages for the image-processing after the images are captured. Recent advances in the GPU-accelerated systems are well-suited for building a performant compute infrastructure for processing CryoEM data.
Understanding CryoEM storage and compute needs
A typical CryoEM run may capture a few thousand images, grouped as movies, and may generate a few terabytes of data per sample (median size 1.2 TB). These movies must be stored for further processing. Moreover, the processing steps produce a large amount of intermediate data. Once the analysis is completed, the researcher may go back to the intermediate and/or original movies for future analysis. A scalable and cost-effective storage solution is needed for short-term, as well as long-term, storage of these files.
Once the images are generated by the microscope, a multi-step compute process is followed for image processing. Figure 1 below provides a typical workflow for CryoEM image processing. Notice that some of the steps in the workflow can be accelerated using GPU-enabled compute.
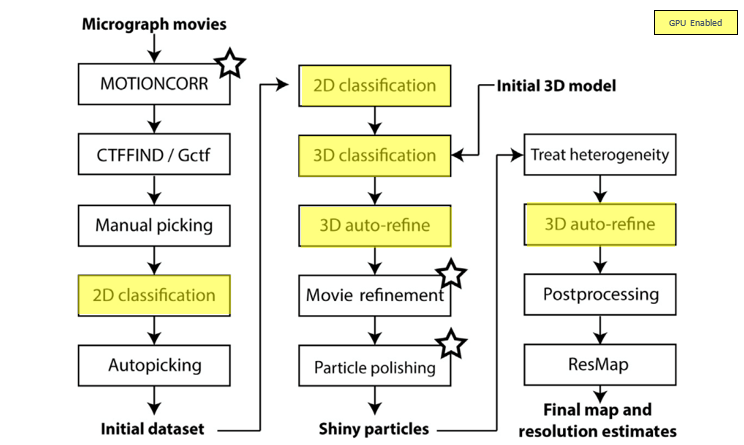
Figure 1: CryoEM image processing – typical workflow
Setting the stage for scalable CryoEM processing
The scaling of CryoEM processing through cloud poses unique opportunities to improve management and processing of large amounts of data and provide scalable compute for reducing computational time and investment necessary for CryoEM structure determination. This facilitates wider adoption of CryoEM as a standard tool for structural biology.
- Data management and transfer
The data volumes generated by the CryoEM workflows are in the TB range per experiment. This data must be made available to the compute workflows in a timely manner. AWS provides a comprehensive portfolio of high-performance transfer and migration options. These include AWS DataSync, AWS Direct Connect or AWS Storage Gateway. You can choose from multiple options to fit your needs. The data can be transferred to an S3 bucket or an EFS volume based on your needs. - Data availability for workflow (S3 vs EBS vs EFS vs Amazon FSx)
As the CryoEM data is needed across multiple workflow steps, the data availability must be carefully designed for cost and throughput optimization. The source data can be stored on an S3 bucket. AWS offers flexible object, block, or file storage for your transient and permanent storage requirements. Depending upon the nature of analysis, the source data from S3 can be copied over to an EBS volume (single EC2 instance using the data), EFS (long-running storage of data to make available for multiple workflow steps simultaneously) or Amazon FSx (transient data available for multiple workflow steps simultaneously). Once the analysis is completed, the data can be stored in long-term storage through lifecycle management. Lifecycle management policies can be attached to the data for moving to low-cost storage (Amazon S3 Glacier, Amazon S3 Glacier Deep Archive, and/or S3 Intelligent-tiering). - Compute and networking – GPU accelerated compute
AWS offers the broadest and deepest platform choice for compute environments. These include general purpose, compute intensive, memory intensive, storage intensive, and GPU compute and graphics intensive. There are 200+ instance types for virtually every workload and business need.As seen in the previous figure (Figure 1), many of the workflow steps can be accelerated using GPU enabled EC2 instances. AWS delivers proven, high performance GPU-based parallel cloud infrastructure to provide every developer and data scientist with the most sophisticated compute resources available today. AWS is the world’s first cloud provider to offer NVIDIA® Tesla® V100 GPUs with Amazon EC2 P3 instances, which are optimized for compute-intensive workloads. The following table lists the CryoEM workflow relevant GPU instance types available today. More details available at https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/accelerated-computing-instances.html
Figure 3: Accelerated computing EC2 instances for CryoEM
You can also avail these instances as Spot Instances for reducing costs further. A huge benefit of the AWS Cloud is that these expensive resources only occur costs when they are in use, greatly saving costs compared to data center installations.
- Automation and orchestration
Generally, it is preferred to use managed and serverless infrastructure. The CryoEM workflow is well-suited for building automated, secure, and resilient workflow using AWS Batch and AWS Step Functions, while reducing the amount of code and infrastructure that you have to write/create and maintain.With the use of AWS Step Functions, you can automate the processing of the data as a multi-step process, while adding any needed functionality by leveraging the ability to integrate with other AWS services such as AWS Lambda, AWS Batch, Amazon DynamoDB, Amazon ECS, Amazon SNS, Amazon SQS, and AWS Glue.The algorithms used in analysis steps can be containerized using Docker and integrated with Amazon ECS – providing consistency and fidelity, distributed compute, scalability, and ability to efficiently use available system resources. - AWS Batch and integrated support for EC2 accelerated instances
AWS Batch enables developers, scientists, and engineers to easily and efficiently run a large number of batch computing jobs on AWS. It dynamically provisions the optimal type and quantity of compute resources based on volume and specific resource requirements. AWS Batch has integrated support for GPU-enabled EC2 instances. This allows for building complex compute environments seamlessly for CPU/GPU based hybrid workloads like CryoEM. - Operation and management
Amazon CloudWatch can be integrated into above described workflow by obtaining data and actionable insights to monitor your applications, understand, and respond to any failures and performance changes, and in general get a unified view of operational health as well as workflow status.
Implementing the workflow
The diagram below provides a typical reference architecture for running the CryoEM workflow.

Figure 4: Typical Reference Architecture for building CryoEM workflow
The Cryo-electron microscope produces images of the sample in question, and the image files are loaded to an S3 bucket. The data stored on an S3 bucket can be copied over onto a POSIX file system such as Amazon EBS, Amazon FSx or Amazon EFS that can be mounted on an EC2 instance based on the data availability needs.
A workflow configuration is typically built using Amazon States Language (ASL) that can be interpreted by AWS Step Functions. The workflow configuration file contains the steps used in processing the files, including the type of compute environment (GPU, vCPU, memory), algorithm container image used in analysis, the commands used for processing individual steps, and any additional parameters such as MPI-threads. It may also contain mount-point binding for the Docker containers, along with any additional requirements. AWS Step Functions use the workflow configuration file to orchestrate the entire workflow for image processing. AWS Step Functions sit on top of AWS Batch and allow you to decouple the workflow definition from the execution of individual jobs.
AWS Batch orchestrates the individual job, and dynamically provisions the optimal quantity and type of compute resources based on the volumes and specific resource requirements of the batch job submitted. AWS Batch plans, schedules, and executes these workloads across the full range of AWS compute services and features such as EC2 and Spot Instances.
Jobs are the unit of work executed by AWS Batch. Jobs can be executed as containerized applications running on Amazon ECS task container instances in an ECS cluster. Once the job is completed, the output of the job can be stored on an S3 bucket. Additionally, workflows that you create with AWS Step Functions can connect and coordinate other AWS services including running an Amazon ECS, submission of the job to a follow-on AWS Batch job. Once all the jobs are successfully completed, the results can be pushed to an S3 bucket for further evaluation. The job progression can be monitored using CloudWatch Logs.
Final thoughts
Access to streamlined computational resources remains a significant bottleneck for new users of cryo-electron microscopy (CryoEM). To address this, we have developed an architecture for building the infrastructure as well as running the CryoEM analysis routines and model building jobs on AWS.
This architecture allows users to move data to the cloud, incorporate appropriate security, and cost-saving strategies, giving novice users access to complex CryoEM data processing pipelines. This architecture dramatically reduces the barrier for entry of new users to cloud computing for CryoEM, and allows them to focus on accelerated scientific discovery.
Interested in implementing a GPU-enabled CryoEM workflow? Contact your account manager or contact us here to get started. Also, keep an eye out on the AWS for Industries blog for our follow-up post that will step through workflow implementation in detail.