AWS for Industries
Creating an Interoperable Clinical Voice Application with AWS
In this blog we will highlight a presentation we gave at re:Invent 2021: Creating interoperable real-time clinical applications. We will demonstrate how to create a clinical voice driven application using Amazon HealthLake to store data in Fast Healthcare Interoperability Resources (FHIR) R4. Amazon HealthLake is a HIPAA-eligible service offering healthcare and life sciences companies a complete view of individual or patient population health data for query and analytics at scale.
Before Getting Started
Healthcare organizations utilize a variety of solutions such as Electronic Health Records (EHR’s), patient portals, telehealth, digital front door solutions, and department specific solutions. These numerous applications often have their own databases and different data models, thus creating silos of data within the organization. We need to avoid creating data silos when creating a new real-time clinical application. On AWS, you can create a central location of your clinical data, where you then build cloud-centric healthcare applications, new AI/Machine Learning (ML) models, and dashboards with the actual data―not copies of data―by using common data models in one safe and secure place.
Utilizing a centralized location for your clinical data can have many advantages, such as:
- A single source of truth for your clinical data.
- Avoiding data duplication,
- the risk of data drift, and
- inaccuracies caused by conflicting data sources not kept up-to-date.
- Simplified security and compliance for your clinical data storage.
- Applications should be architected with security and privacy designed-in, not bolted on at the end.
- This is critical to ensuring that you are building a solution which meets all of your regulatory and compliance requirements up front, while being maintainable and observable.
- Applications should be architected with security and privacy designed-in, not bolted on at the end.
Overview of solution
For creating a voice solution, the key AWS services we will focus on are:
- Amazon Cognito for authentication
- Amazon Simple Storage Service (Amazon S3) for storing audio files
- Amazon Transcribe Medical for medical speech to text
- Amazon HealthLake (HealthLake) to store our data in the Fast Healthcare and Interoperability Resources (FHIR) R4 format
- AWS Lambda (Lambda) for serverless execution of application specific code
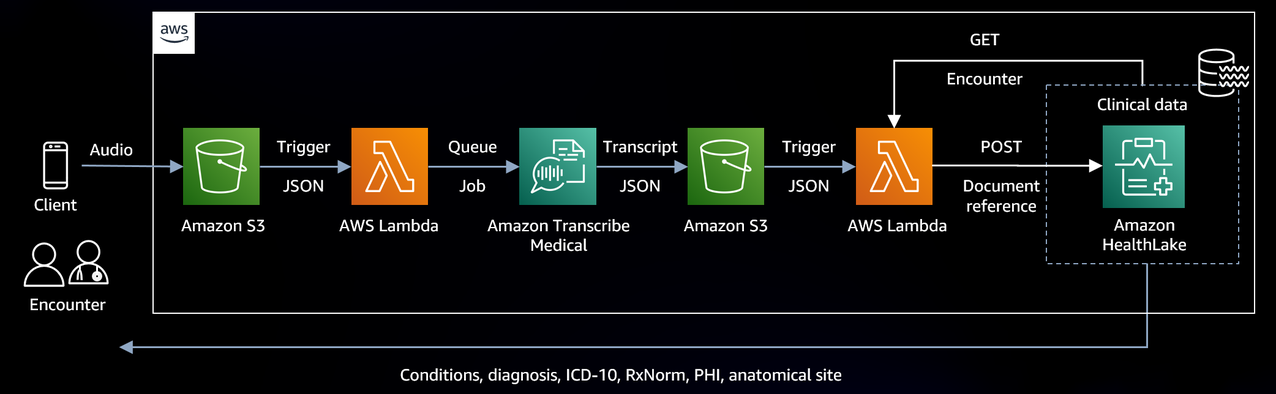
We will run AI/ML medical models against our conversation to surface results back to clinicians. This will give us a common data model to build with, and store our data in a centralized place, which can be accessed by other AWS services in our account. Below is the high-level architecture we will be following.

Figure 1 – High-level AWS interoperable audio architecture overview
Prerequisites
For this walkthrough, you should have the following prerequisites:
- An AWS account
- Ability to create AWS resources and assign AWS Identity and Access Management (IAM) roles
- General understanding of FHIR and RESTful API’s
- A HealthLake store that you can write FHIR resources to
Walkthrough
Let’s first dive into what the data flow looks like. Also, note that while we’re focused on audio, the architecture and workflow we are outlining can support other media types by simply swapping Amazon Transcribe Medical for other AWS AI/ML services, such as Amazon Textract for images and documents. That said, everything starts with the encounter. For this solution, we’re allowing the audio to be captured and uploaded to an Amazon S3 bucket. Amazon Transcribe Medical is also designed to support real-time streaming as well. However, we wanted to create a solution that allows us to load historic and near real-time audio files that may have been captured while offline.
Figure 2 – Voice to HealthLake data store process flow
Step 1: Convert Clinical Speech to Text
The above architecture illustrates an Amazon S3 trigger to run a Lambda function when a new audio object is created. The audio object is passed to Amazon Transcribe Medical to be converted into text using specialized natural language understanding AI models to accurately transcribe medical terminologies such as medicine names, procedures, and even conditions or diseases to text.
Create a Lambda function that will be triggered upon audio object creation in your Amazon S3 bucket when you load new audio files to it. The example of a Python Lambda handler function (Python 3.7) below takes as input an Amazon S3 event and initiates an asynchronous transcription request using the Amazon Transcribe Medical API. The results of the transcription process will be saved to the Amazon S3 bucket specified in the “OutputBucketName” parameter.
import boto3
import uuid
import json
def lambda_handler(event, context):
record = event['Records'][0]
s3bucket = record['s3']['bucket']['name']
s3object = record['s3']['object']['key']
s3Path = "s3://" + s3bucket + "/" + s3object
jobName = s3object + '-' + str(uuid.uuid4())
client = boto3.client('transcribe')
response = client.start_medical_transcription_job(
MedicalTranscriptionJobName=jobName,
LanguageCode='en-US',
MediaFormat='mp4',
Media={
'MediaFileUri': s3Path
},
OutputBucketName="transcribe-output-demo",
Settings={
'ShowSpeakerLabels': True,
'MaxSpeakerLabels': 2,
'ChannelIdentification': False,
'ShowAlternatives': False
},
ContentIdentificationType='PHI',
Specialty='PRIMARYCARE',
Type='CONVERSATION'
)
return {
'TranscriptionJobName': response['MedicalTranscriptionJob']['MedicalTranscriptionJobName']
}Step 2: Create the FHIR DocumentReference
When Amazon Transcribe Medical completes the speech to text processing and saves the output to the target Amazon S3 bucket, an Amazon S3 trigger is again used to call a Lambda function. The text that is generated by Amazon Transcribe Medical will need to be stored in a JSON structure representing a FHIR DocumentReference.
A DocumentReference is: a resource used to index a document, clinical note, and other binary objects such as a photo, video, or audio recording, including those resulting from diagnostic or care provision procedures, to make them available to a healthcare system. A document is some sequence of bytes that is identifiable, establishes its own context (an example, what subject, or author can be presented to the user), and has defined update management. The DocumentReference resource can be used with any document format that has a recognized mime type and that conforms to this definition.
The below Python code shows how a basic DocumentReference structure can be created. One item to note is the text of the actual document is expected to be base64 encoded. The function below takes several parameters to represent the unique instance attributes of a specific document instance and will be customized based on your solution’s specific needs. Create a secondary Lambda function to trigger from the Amazon S3 bucket (which receives the transcription results) and call this function in it.
def createDocRef( docText, transcriptionTime, subject, idEncounter, encounterStartTime, encounterEndTime, serviceProvider, serviceProviderDisplay, practID, practDisplay ):
encodedText = base64. \
b64encode( str(docText).encode("ascii") ). \
decode('utf-8')
jsonDocRefTemplate =
{
"resourceType":"DocumentReference",
"date":"transcriptionTime",
"custodian":{
"reference":"serviceProvider",
"display":"serviceProviderDisplay"
},
"subject":{
"reference":"subject"
},
"author":[
{
"reference":"practID",
"display":"practDisplay"
}
],
"context":{
"period":{
"start":"encounterStartTime",
"end":"encounterEndTime"
},
"encounter":[
{
"reference":"idEncounter"
}
]
},
"type":{
"coding":[
{
"system":"http://loinc.org",
"code":"75519-9",
"display":"Encounter"
}
]
},
"category":[
{
"coding":[
{
"system":"http://hl7.org/fhir/us/core/CodeSystem/us-core-documentreference-category",
"code":"clinical-note",
"display":"Clinical Note"
}
]
}
],
"content":[
{
"attachment":{
"data":"encodedText",
"contentType":"text/plain"
}
}
],
"status":"superseded"
}
return( jsonDocRefTemplate )Once this object is created in your code, it can be stored in HealthLake.
Step 3: Store the DocumentReference in Amazon HealthLake
Ensure your Lambda function or other executable resource has a permission policy that enables creating resources in the target HealthLake datastore. For example, the below IAM policy allows for the creation of resources, “healthlake:CreateResource”, in the target HealthLake store, “arn:aws:healthlake:us-east-1:<account>:datastore/fhir/<datastore id>” where accountid and datastoreid represent the specific instances within your account. Identity and Access Management for Amazon HealthLake provides a detailed discussion on access management with Amazon HealthLake.
{
"Version": "2012-10-17",
"Statement":
[
{
"Sid": "VisualEditor0",
"Effect": "Allow",
"Action": "healthlake:CreateResource",
"Resource": "arn:aws:healthlake:us-east-1: 555555555555 :datastore/fhir/1234567890abcdef01234567890abcde"
}
]
}To create the DocumentReference in HealthLake, a standard FHIR REST API will be used to create the new resource. We use the POST request type and the payload of the request containing the JSON representing the new DocumentReference object as shown in the below Python code snippet.
# Replace region and healthlake_url with your solutions method for specifying these values
service = 'healthlake'
region = 'us-east-1'
datastoreid = '1234567890abcdef01234567890abcde'
healthlake_url = f"https://{service}.{region}.amazonaws.com/datastore/{datastoreid}/r4/"
headershl = {
'Host': f"{service}.{region}.amazonaws.com", 'Content-Type': 'application/json'
}
# Create the DocumentReference with encounter specific parameters
jsonDocRef = createDocRef( docText, transcriptionTime, subject, encounter, encounterStartTime, encounterEndTime, serviceProvider, serviceProviderDisplay, practID, practDisplay)
# POST to HealthLake
hldocrefendpoint = healthlake_url + 'DocumentReference'
ipayload = json.dumps(jsonDocRef)
request = AWSRequest(method='POST', url=hldocrefendpoint, data=ipayload, headers=headershl)
SigV4Auth(boto3.Session().get_credentials(), service, region).add_auth(request)
session = URLLib3Session()
request_result = session.send(request.prepare())In your specific implementation of this code snippet, the value of region and “healthlake_url” will need to be initialized based on your specific solution’s method for maintaining the instance specific values. In this example, the complete endpoint URL to create a new document reference would be:
https://healthlake.us-east-1.amazonaws.com/datastore/1234567890abcdef01234567890abcde /r4/DocumentReference
Upon successful completion of the create resource operation that is implemented as a POST request to the Amazon HealthLake datastore endpoint, the HTTP response code will be ‘201’, indicating the request successfully created the resource. The body of the response will contain the new resource with additional common FHIR parameters created. These new attributes are the resource “id” and the meta tag value “lastUpdated”. The resource id uniquely identifies the new resource in the FHIR repository. The lastUpdated attribute represents the last time the resource was modified, in this case, when it was created. An example of a returned document reference is below, with the FHIR server created values at the bottom.
{
"resourceType":"DocumentReference",
"date":"2021-10-22T13:51:49-08:00",
"custodian":{
"reference":"Organization/6e21d2e1-b52e-38fa-8254-8a797651c2d0",
"display":"PCP8941"
},
"subject":{
"reference":"Patient/e50bd311-353f-46c3-a25b-b8fcd957735d"
},
"author":[
{
"reference":"Practitioner/f4a76de4-a0fd-34ca-8b04-2935b1270702",
"display":"Dr. Kory651 Fisher429"
}
],
"context":{
"period":{
"start":"2021-10-22T13:36:49-08:00",
"end":"2021-10-22T13:51:49-08:00"
},
"encounter":[
{
"reference":"encounter/d8350610-c1ac-4b09-b4c4-63ca3a6bcb7d"
}
]
},
"type":{
"coding":[
{
"system":"http://loinc.org",
"code":"34117-2",
"display":"History and physical note"
},
{
"system":"http://loinc.org",
"code":"51847-2",
"display":"Evaluation+Plan note"
}
]
},
"category":[
{
"coding":[
{
"system":"http://hl7.org/fhir/us/core/CodeSystem/us-core-documentreference-category",
"code":"clinical-note",
"display":"Clinical Note"
}
]
}
],
"content":[
{
"attachment":{
"data":"SGksIEJ...aHQgbm93Lg==",
"contentType":"text/plain"
}
}
],
"status":"superseded",
"id":"85fbb539-16e5-4d34-a60c-488de44b813e",
"meta":{
"lastUpdated":"2021-10-01T20:37:56.121Z"
}
}Step 4: Integrated use of Amazon Comprehend Medical augments the DocumentReference
Lastly, Amazon HealthLake’s integrated use of Amazon Comprehend Medical extracts the medical terminology and augments the DocumentReference resources with attributes it discovers when a new DocumentResource is created. These attributes (conditions, diagnosis, ICD and RxNorm codes, anatomical sites, and other information) can then be surfaced back to the clinician by the application. These new attributes are automatically appended to the DocumentReference in an extension attribute and are fully described in the HealthLake – Integrated medical natural language processing documentation, including complete example outputs.
A condensed snippet of the JSON resource extension is below (Note: “….” represent areas that were omitted for readability):
"extension":[
{
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/raw-response",
"valueString":"{Entities: [{Id: 0,Text: asthma,Category: MEDICAL_CONDITION,Type: DX_NAME,Score: 0.98681927,BeginOffset: 143,EndOffset: 149,Attributes: [],Traits: [{Name: DIAGNOSIS,Score: 0.73062783}],ICD10CMConcepts: [{Description: Unspecified asthma, uncomplicated,Code: J45.909,Score: 0.8120982}, {Description: Unspecified asthma,Code: J45.90,Score: 0.720394}, {Description: Unspecified asthma with (acute) exacerbation,Code: J45.901,Score: 0.7050891}, {Description: Other asthma,Code: J45.998,Score: 0.68928576}, .... ,ModelVersion: 0.1.0}"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/model-version",
"valueString":"0.1.0"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-id",
"valueInteger":0
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-text",
"valueString":"asthma"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-begin-offset",
"valueInteger":143
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-end-offset",
"valueInteger":149
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-score",
"valueDecimal":0.98681927
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-ConceptList",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Code",
"valueString":"J45.909"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Description",
"valueString":"Unspecified asthma, uncomplicated"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Score",
"valueDecimal":0.8120982
}
]
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Code",
"valueString":"J45.90"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Description",
"valueString":"Unspecified asthma"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Score",
"valueDecimal":0.720394
}
]
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Code",
"valueString":"J45.901"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Description",
"valueString":"Unspecified asthma with (acute) exacerbation"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Score",
"valueDecimal":0.7050891
}
]
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Code",
"valueString":"J45.998"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Description",
"valueString":"Other asthma"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Score",
"valueDecimal":0.68928576
}
]
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept",
"extension":[
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Code",
"valueString":"J45.9"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Description",
"valueString":"Other and unspecified asthma"
},
{
"url":"http://healthlake.amazonaws.com/aws-cm/infer-icd10/aws-cm-icd10-entity-Concept-Score",
"valueDecimal":0.6697552
}
]
}
]
}
]
},
"......."In the above example you saw sets of the ICD-10 coding inferred from the document text which was generated from the audio that was captured. For each inference, there is also an entity-score representing the underling model’s confidence score of the result. This score can be used to filter results and determine which should be presented to clinicians for confirmation during a complete workflow.
Conclusion
The described architecture and concepts in this blog can be applied to various use cases and be augmented with other input modalities based on those use case’s unique requirements. Example use cases for audio transcription could include:
- Ambient listening to doctor patient engagements to provide clinical note taking based on the transcribed audio and the augmented DocumentReference inferences
- Clinical rounds transcribed from recorded audio and then reviewed by the clinician
- Replace Amazon Transcribe Medical with Amazon Textract and process written clinical notes in the same manner
We encourage you to reach out to your AWS account teams and solution architects and work with them to evaluate how Amazon Transcribe Medical and HealthLake can be used to satisfy your own clinical solution needs. Using the breadth of AWS services, your teams can realize the benefits of breaking down your data silos to achieve a single source of truth for your clinical data and simplifying your overall security and compliance posture. By leveraging AWS services to provide the undifferentiated heavy lifting in your applications, you can improve the overall patient experience by creating solutions that allow clinicians more time with their patients, and not the computer.
As noted earlier, this entire solution was presented by us at re:Invent 2021. You can view our presentation, as well as a working demo of this solution, by viewing: Creating interoperable real-time clinical applications.