AWS for Industries
Predicting all-cause patient readmission risk using AWS data lake and machine learning
It’s no secret that hospital readmissions impact patient outcomes and the financial health of healthcare providers globally. Specifically in the United States, the Agency for Healthcare Research and Quality (AHRQ) shows that readmissions are some of the costliest episodes to treat, with costs reaching in excess of $41.3B. Hospitals and healthcare providers are looking for ways to reduce the preventable hospital readmission to improve the quality of care provided to patients and comply with Affordable Care Act. As a part of the Affordable Care Act, the Hospital Readmissions Reduction Program requires the Centers for Medicare and Medicaid Services (CMS) to penalize hospitals for “excess” levels of readmissions. In 2019, Medicare cut payments to 2583 hospitals costing these institutions over $563 million over the course of a year, impacting the financial health of these organizations.
Data and machine learning as a solution
Healthcare Providers and Payers are identifying innovative ways to predict and prevent hospital readmissions to improve the health of their patients and avoid financial penalties. Large-scale adoption of Electronic Health Record (EHR) system, a standard to maintain healthcare records electronically has created the opportunity to mine data to identify the risk of hospital readmission. The volume, variety, and velocity of the data collected by Healthcare providers make it difficult to not only manage the data but also generate valuable insights from large datasets. Machine learning solutions have proven to be powerful tools to predict patient readmission risk as stated by a number of research studies. A real world example of this being put into practice can be seen here. Chairman & CEO of Cerner Corporation, Brent Shafer, shares how they have transformed patient care by using machine learning technologies to predict hospital readmissions, improving the lives of the patient, family, and caregiver.
Building a hospital readmission prediction system
Architects and Developers throughout the healthcare ecosystem are looking to build a Hospital Readmission Risk Prediction system that will improve patient care, lower the cost of care and ultimately increase the providers Medicare reimbursement. This blog focuses on how to use the provided reference architecture to quickly develop end-to-end machine learning solution for ‘All Cause Hospital Readmission Prediction System’ using large EHR datasets. The purpose of this blog is not to provide machine learning algorithm or datasets for the problem but to show how different AWS services can be used together to build the prediction system. This solution includes data ingestion, data exploration, feature selection, feature engineering, data preprocessing, and training ML models. It also includes performance optimization of the model, deployment of the model as an inference pipeline, and real-time prediction for the provided input dataset. Having access to representative data is essential for any machine learning problem. Since real healthcare data is highly regulated, we are going to use Synthetic Patient Population Simulator (Open Source tool) to generate synthetic and realistic EHR data for the purpose of this blog. This tool provides flexibility to generate datasets for varied population sizes, geographic regions, demographics, and many other configurable items (mentioned in synthea.properties file within the tool). You can refer to the steps mentioned on this page for the latest instructions to download, install, and generate the dataset. In a nutshell, you must:
- Install Java and clone the Git repo
- Verify the installation
- Update src/main/resources/synthea.properties file to generate files in csv format. Ensure that the property exporter.csv.export is set to true (exporter.csv.export = true)
- Run the tool (specifying population size) to generate dataset (For example, ./run_synthea -p 10000). The latest command can be found on this page.
This generates the dataset in “output/csv” directory of the cloned directory structure. Some of the files generated by the tool are imaging_studies.csv, allergies.csv, payer_transitions.csv, patients.csv, encounters.csv, conditions.csv, medications.csv, careplans.csv, observations.csv, procedures.csv, immunizations.csv, organizations.csv, providers.csv, and payers.csv. These files contain standard EHR data. To understand the data dictionary of the generated dataset, you can refer to this link.
For the purpose of this blog, we have used the synthetic data generated by the tool but you have the flexibility to use your own data with this reference architecture and modify the provided scripts to generate training dataset and build machine learning model. Let’s look at the reference architecture.
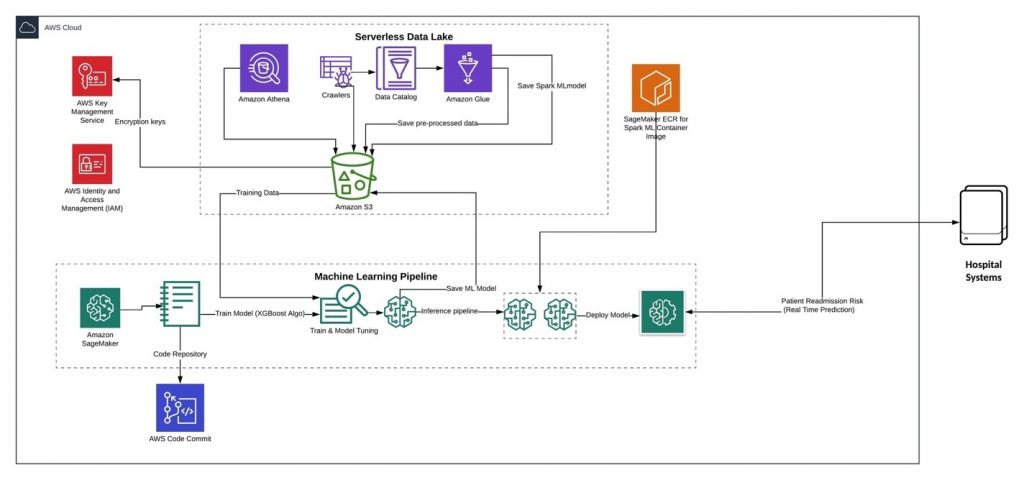
In this reference architecture-
- Amazon S3 is used to store raw datasets, pre-processed training, and test datasets and machine learning model artifacts
- AWS Glue is used to catalog the schema, converting csv into parquet, feature selection, feature engineering and generate Spark machine learning (ML) model for building inference pipeline.
- Amazon Athena to run ad hoc queries to understand the dataset and identify relationship between different values and attributes within the data.
- Amazon SageMaker to provision Jupyter notebook, which is used to train the required machine learning model, model hyper parameter tuning, create SageMaker model artifacts and deploy real-time prediction inference pipeline.
- AWS Key Management Service (KMS) to protect the data at rest and control data access by different AWS services.
Since you need HIPAA compliance for Healthcare data, the architecture in based on AWS HIPAA eligible services. For more details on architecting for HIPAA compliance, please refer to HIPAA on AWS Whitepaper.
Below is the link to the CloudFormation template, which can create the required AWS resources to launch this solution in your AWS Account.
For detailed instructions, please refer to this GitHub Repo
As a summary, the steps that you must follow for creating and deploying machine learning model from the above generated data are as follows:
- Understanding of your data
- Storing and converting your data into Apache Parquet for optimized performance and storage
- Feature selection and engineering using Apache Spark
- Data pre-processing to convert categorical variables into required training data
- Train Spark ML model for data pre-processing and then serialize using MLeap to be used during Inference Pipeline
- Convert the dataset into XGBoost supported format (i.e., CSV from Spark Data Frame)
- Split the dataset into training and validation for model training and validation
- Train XGBoost model using Amazon SageMaker XGBoost algorithm and validate model prediction using validation dataset
- Tune the trained model using Hyperparameter tuning jobs
- Get the best tuned model and create inference pipeline that includes Spark ML model and XGBoost model
- Create the endpoint configuration to deploy the inference pipeline
- Deploy the inference pipeline for real-time prediction
- Invoke real-time prediction API for a request.
Preparing raw data
Check the bucket policy to see that the stack created the bucket policy as below to ensure that no objects on this bucket are unencrypted.
After the data is uploaded, you must run AWS Glue crawler to discover the schema of all uploaded CSV files and update the database with corresponding tables and attributes. CloudFormation stack creates the required Glue crawlers, security configuration and Glue database for that crawler. You can log in to AWS Management Console to run the crawler, look for the crawler named ‘ehr-crawler-readmission’ (default name provided in CloudFormation template) and run the crawler.
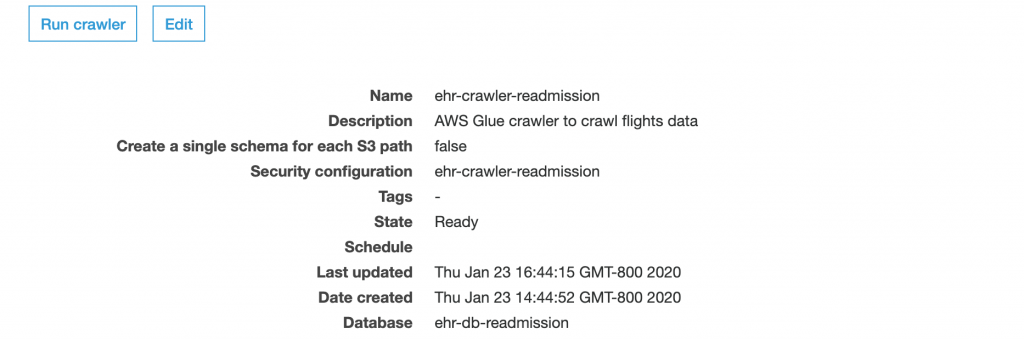
Once the crawler is successfully run, go to databases in AWS Glue console and look for the Glue Database named ‘ehr-db-readmission’(default name provided in CloudFormation template). You can click on the link ‘Tables in ehr-db-readmission’ to check the available tables and associated properties.

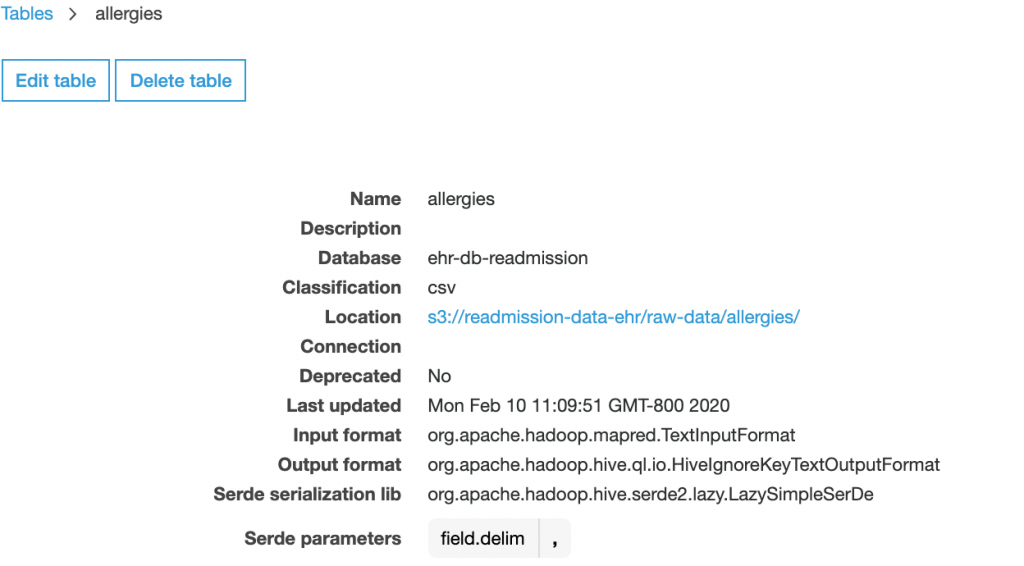
When you click on a table as shown below, you can check the different attributes, input and output format of the files, columns, and data types along with the partition keys.
Feature selection, feature engineering, and data pre-processing using Apache Spark in AWS Glue
Now, you have understood the table structure and attributes. You want to get started with data pre-processing steps. We use two AWS Glue Jobs for this purpose, which are – one for converting CSV files into Parquet for better query performance and storage optimization and second for feature selection, feature engineering and data pre-processing. You can now log in to Amazon SageMaker Console and click on ‘Open Jupyter’ link to open the Jupyter Notebook Instance provided as part of this blog. Further instructions on what must be done are mentioned in the Notebooks. Open the notebook ‘readmission-risk-inference-pipeline-evaluation’ to follow the instructions. If you want to quickly go through the overall process and important pieces of code, you can continue reading this blog.
Let’s look at a few important steps from the notebook to understand them better.
Machine learning (ML) models require data to be available in a specific format to do inference. This would mean to convert categorical columns into ML model-specific format before the request can be passed to classification model for inference. To avoid this as an additional step, you can create data pre-processing model and deploy this model along with your classification model as a single entity for real-time prediction. The below step is required to download the required dependencies for MLeap to serialize Spark ML model for data pre-processing so that we can include this model in SageMaker Inference Pipeline at later stage.
You need PySpark scripts to do transformation and pre-processing on the large datasets. You may want to leverage the benefits of distributed processing for better performance and scalability. You can look at the scripts in detail by going to AWS Glue Job console.
First, you must convert CSV files into Parquet using the below AWS Glue Job. You can generate the scripts in AWS Glue using the options in the console so that you don’t have to write the script from scratch and can make modifications to the generated script as per your use case. In this case, the script is provided to you to read the data from AWS Glue Crawler Database and filter the columns. The script drops the null values and update the data types to as supported by the machine learning algorithm. Henceforth, the partitioned dataset is saved to S3 bucket in Parquet format.
After the data is converted to required format, you need to another AWS Glue Job that will produce the training data required for the Amazon SageMaker XGBoost model. This script read data directly from all partitions of the data on S3 bucket but you can filter to read specific partitions. Since you are using Supervised learning model i.e. XGBoost we must provide the label data. You calculate label data, in this case, 30-day readmission for a patient. Once feature selection and engineering is done, you generate the features vector by using Spark ML OneHotEncoding and then finally serialize the model using MLeap library. XGBoost algorithm supports CSV format for training data so you must convert Spark Data Frame into CSV files and save to S3 bucket for training input.
Note – This script is run on Spark 2.2 since MLeap libraries currently support Spark 2.2. You can check more details on latest versions on https://github.com/aws/sagemaker-sparkml-serving-container.
Train and build the Amazon SageMaker XGBoost model
You are going to use Amazon SageMaker XGBoost framework to train your model. Since this is a binary classification problem, you can use binary logistic for training model and AUC curve as objective function for this model. You can also use any other learning task for XGBoost or choose other algorithms that are best suited for your use case and dataset. You have to specify the location of training and validation dataset, EC2 instance configuration, and number of instances for distributed training option to reduce the overall training time. You can also choose to use Spot Instances on SageMaker for training, which can significantly reduce training cost. Once the model is trained, you tune the model performance through hyperparameter optimization. Hyperparameter tuning on SageMaker provides two ways to create tuning jobs, either using Bayesian Search or Random Search. The tuning runs multiple training jobs with different hyperparameter values from the specified range and identify the model that has the best value for objective function.
Deploy the Amazon SageMaker Inference pipeline for real-time prediction
Now you have the model artifacts, you can setup SageMaker Inference Pipeline to combine pre-processing, predictions and post-processing data science tasks into a single model. Note that data pre-processing model is based on Apache Spark ML and classification model is based on SageMaker XGBoost algorithm, which means that you must specify two different containers images while creating the definition of pipeline model. The combination of these two container images along with the model artifacts is deployed behind a single API endpoint in the specified order. Once you have created the pipeline model, you create the endpoint configuration that dictates the instance configuration in which you have the flexibility to specify auto-scaling behavior for the production variant, along with other best practices for security and resilience. You can now use this inference pipeline to make real-time predictions directly without performing external data pre-processing. The code for this step is as below –
Conclusion
Congratulations! You have now learned how to develop real-time prediction system for all-Cause Hospital Readmission using machine learning and serverless technologies. In this process, you learned how to create the required AWS services for end-to-end solution using CloudFormation template. You also learned how to automatically discover unstructured data on Amazon S3 using crawlers in AWS Glue, pre-processing large datasets using Apache Spark in AWS Glue, partitioning of data on Amazon S3, ways to encrypt data at rest using AWS KMS and data in transit using TLS/SSL to protect your data, train model on identified features using training jobs in AWS SageMaker and create inference pipeline for real-time prediction using the Amazon SageMaker Inference Pipeline.