AWS for Industries
Running WDL workflows at scale with Amazon Genomics CLI and MiniWDL
Today, AWS for Genomics announces the addition of miniWDL as a workflow engine in Amazon Genomics CLI, an open source tool that makes it easier to run genomics workflows at-scale by automating the deployment of workflow engines and compute clusters. MiniWDL is a lightweight engine for running Workflow Definition Language (WDL) based workflows developed by the Chan Zuckerberg Initiative (CZI). Using miniWDL with Amazon Genomics CLI provides a highly available and scalable way to run WDL workflows in AWS.
One of the ways Amazon Genomics CLI launches workflow engines is an architecture called “batch-squared”. This serverless method uses a containerized workflow engine “head” in an AWS Batch job per workflow execution. Each workflow “head” job then independently schedules workflow tasks to AWS Batch. In short, AWS Batch handles all of the compute scaling necessary to run both the workflow engine and workflow tasks. This method is a simple solution for running CLI based workflow engines that offers high-availability and is capable of scaling to 1000s of concurrent jobs.

High level view of the “Batch-squared” architecture where each workflow execution has a dedicated engine “head” as an AWS Batch job that submits task jobs to AWS Batch.
MiniWDL is a CLI based WDL engine that uses distinct “head” processes per workflow execution. There is also a new miniWDL plugin that enables miniWDL to submit workflow tasks directly to AWS Batch. Thus, it is now possible to launch miniWDL with “batch-squared” architecture via Amazon Genomics CLI, providing a serverless and highly available and scalable option to run WDL workflows. This looks like:
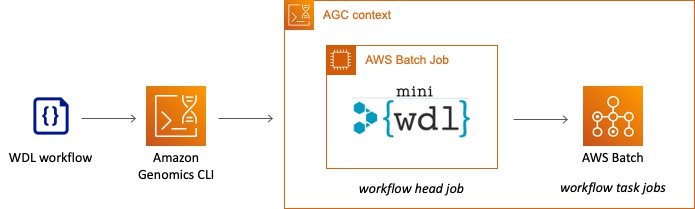
High level view of the running WDL workflows using miniWDL and Amazon Genomics CLI.
To start using miniWDL with Amazon Genomics CLI, first make sure you have Amazon Genomics CLI version 1.2.x (or higher) installed, which you can get from our GitHub repo. You can then configure a context that uses miniWDL by adding the following to your agc-project.yaml config file:
miniwdlCtx:
engines:
- type: wdl
engine: miniwdl
You can then launch this context with:
agc context deploy -c miniwdlCtxOnce launched, you should be able to run all the WDL examples Amazon Genomics CLI provides.
MiniWDL is an official implementation of the Open WDL specification and supports up to WDL 1.1 as of version 1.2.0. Amazon Genomics CLI uses miniWDL version 1.3.x. To date, miniWDL has been a capable WDL engine for local pipeline testing and use cases such as infectious disease metagenomics in CZI’s IDseq platform. MiniWDL’s new plugin for AWS Batch and integration with Amazon Genomics CLI expands its capabilities further, and in our testing it has scaled to meet the needs of whole human genome data processing with typical secondary analysis and joint-genotyping workflows.
Conclusion
The addition of miniWDL support to Amazon Genomics CLI expands the ways customers have to run WDL workflows to include a serverless, easy to use, highly available, and highly scalable option. Overall, the addition of more workflow engines supported by Amazon Genomics CLI allows customers to select the right tooling that meets their performance and cost needs.
To learn more about Amazon Genomics CLI visit our GitHub page, chat with the community on our Gitter channel, and if you have ideas for improvements please send us a pull request!