AWS Startups Blog
How Polly’s Curated Biomedical Molecular Data Streamlines MLOps for Drug Discovery

By Swetabh Pathak, Elucidata founder, and Sahil Rai, Engineering Manager at Elucidata
The application of machine learning to biomedical sciences is crucial to attain success in drug discovery. Biomedical data is large and unstructured, rendering it difficult for ML applications. Equally, many human diseases involve several variables that change dynamically and differ from patient to patient. Understanding this complexity requires unlocking large-scale heterogeneous biomedical and patient data. Machine learning, therefore, can help bioinformatics scientists and drug researchers identify meaningful patterns from large datasets that can lead to better odds of success in the clinic.
Advancements in technologies like next-generation sequencing and mass spectrometry have led to an abundance of data. However, the utilization of this data is still challenging as most of it is not well-curated. The state-of-the-art machine learning models that get better with more data require a viable level of data quality. In sharp contrast, today, most of the biomedical molecular data is available in semi-structured and unstructured formats. When developing new drugs for novel diseases, a faulty model can lead to completely off-the-mark outcomes which are only understood much later in the process. This is where platforms such as Elucidata’s Polly come in handy.
Polly is a DataOps platform that allows data scientists to access ML-ready data generated from data repositories, proprietary experiments, and publications. Curating data into a high-quality resource is the core value proposition of Polly. Polly complements the burgeoning data-centric AI movement that seeks to employ data as a tool to improve ML outcomes.

Using Polly, scientists can manage the life cycle of biomedical molecular data end to end. Polly’s capabilities include:
- Curating data from proprietary, public, and premium sources using active learning to ensure that data is machine-actionable and analysis-ready.
- Storing large volumes of heterogeneous data in one central location.
- Leveraging powerful querying capabilities across dataset, sample, and feature level metadata through SQL.
- Analyzing and visualizing data with the help of a scalable computational platform.
- A toolbox that allows users to either utilize pre-existing bioinformatics pipelines or create custom ones.
- Sharing analysis and results with internal and external collaborators.
Use cases of biomedical molecular data are atypical of other domains where machine learning is used. Data is typically large in volume and low in velocity. Formats are heterogeneous depending upon the kind of data. There are few common metadata standards and crucial metadata is human-generated. Search and discovery of data is hard when data is present in multiple sources, is siloed, and is not labeled consistently. A consistent querying interface is often not available. Polly solves these challenges by incorporating DataOps methodologies. Powered by AWS, Polly’s infrastructure is fast, secure, and scales seamlessly.
How Polly works

AWS has a robust lineup of services that serve a platform’s DataOps demands such as building and managing data pipelines, ML pipelines, and hosting applications at scale with great ease. Four critical workstreams are deployed on AWS:
- Ingestion & ETL infrastructure
- Storage & querying infrastructure
- Data curation infrastructure
- Scalable compute infrastructure
Workstream 1 – Ingestion & ETL infrastructure

Polly ingests and processes hundreds of thousands of datasets every week with an infrastructure capable of scaling to millions of datasets. Polly’s infrastructure is designed to be fault-tolerant and cost-effective. Polly’s ETL infrastructure uses Amazon EC2 Spot instances to reduce the cost of data ingestion by almost 90% compared to using regular on-demand instances. The steps involved in Polly’s ETL pipelines are:
- Datasets from multiple sources are added to an Amazon Simple Storage Service (Amazon S3) bucket. There is an S3-to-lambda trigger set up on every dataset that carries out the upload/delete/update function. Next, the dataset ID is sent to the Controller lambda.
- The Controller lambda adds a row in the Amazon DynamoDB table and assigns a job ID. The status of the job is marked as “in-queue”. DynamoDB is used to store Job ID, job metadata, and job state. (Note: Each dataset update is considered as one job).
- After adding an item in the DynamoDB, the Controller lambda sends a message to Amazon Simple Queue Service (Amazon SQS). The message contains the job ID.
- Messages in the SQS trigger the Scaling lambda.
- Scaling lambda starts scaling out the ECS Cluster (spot fleet), which fetches the job metadata based on the job ID.
- These ECS clusters transform and load the datasets to Amazon OpenSearch and in S3.
- Once loaded, the status of jobs is updated on the DynamoDB table (as Success/Errored).
- The time-based Amazon CloudWatch event rule keeps triggering the scaling lambda. Scaling lambda keeps on checking “Approximate number of messages in the queue”. In case of no messages, scaling lambda starts scaling in the spot instances. The last three steps are repeated until there are no messages in the queue.
Workstream 2 – Storage & querying infrastructure
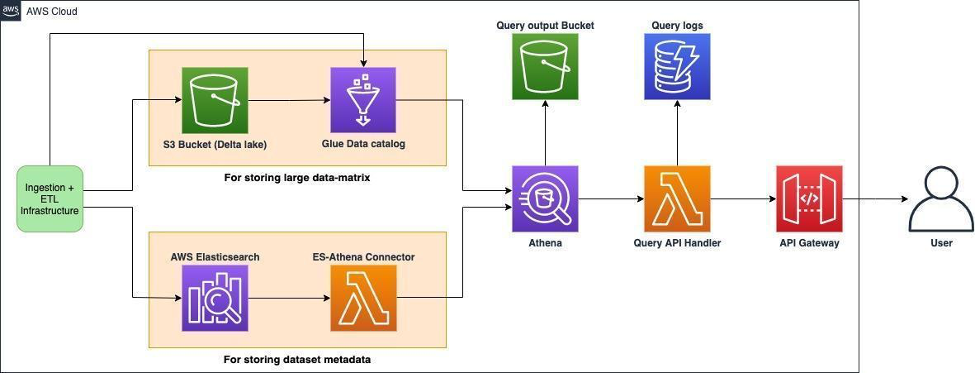
Once the datasets are ingested, the goal is to be able to query and search them effectively. A dataset has two major components: metadata and data matrices. The schema of metadata is semi-structured. On the other hand, data matrices are structured entities, mainly containing numerical values. Search is facilitated by the metadata of the datasets. Some searching algorithms also utilize the underlying data matrices. Hence metadata and data matrices are stored in the Amazon OpenSearch service and Amazon S3 bucket (as Delta Lake) respectively. One challenge with this approach is that it is difficult to query two different sources separately and then combine the results. That’s where Amazon Athena comes into play.
Amazon Athena lets federated queries run on two different sources using SQL syntax, delivering a unified interface to our users. To do this, we have used the following strategy:
- The ETL infrastructure loads datasets into Amazon Elasticsearch and S3 bucket. The data in S3 is stored as a Delta Lake.
- Elasticsearch is used to store datasets with diverse metadata such as types of diseases, drugs, organisms, etc.
- Delta Lake is used to store large 2D data matrices, which contain numerical data. Delta Lake enables us to store and query on-disk data using ACID transactions.
- Amazon Athena is then used to query both Elasticsearch and S3 bucket. Querying Elasticsearch is facilitated by the ES-Athena connector. A Glue data catalog is used to store the schema of the data present in Delta Lake.
- API Gateway is used in combination with AWS Lambda to proxy the Athena endpoint. Having a custom API layer over Athena ensures that users have consistent access to all Polly’s services.
Workstream 3 – Data curation infrastructure
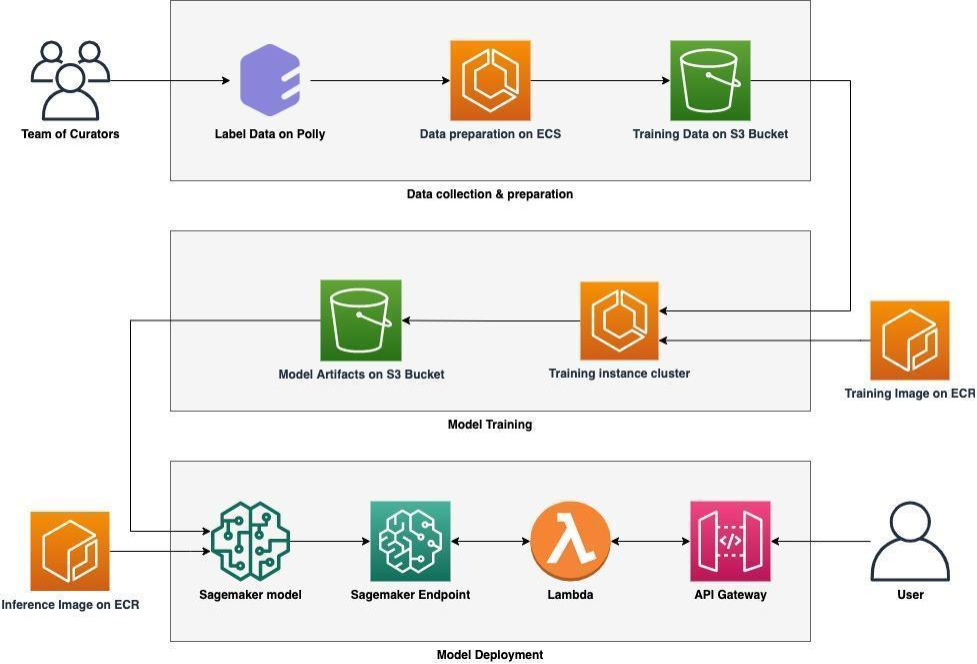
The metadata quality of a dataset is improved by adding additional fields and filling the gaps in existing fields. This is done by our proprietary ML framework called PollyBERT. PollyBERT has been trained on millions of labels generated by our team of experts who label the datasets on our expert curation platform. The training data is used to scale the annotations to millions of datasets using multiple ML models.
Operating this ML pipeline can be a daunting task. This is why we have designed and developed an end-to-end machine learning pipeline using AWS services. This pipeline consists of 3 sequential steps starting with data collection and preparation, followed by model training, and finally deployment and hosting of the inference model using Sagemaker.
Polly’s data labeling infrastructure has 3 major components:
1) Collecting Training Data
- Data labeling happens on Polly’s expert curation infra by expert annotators. For example, an annotator might label a dataset with the associated disease or drug.
- After receiving the labeled data, the data is pre-processed using Amazon ECS clusters and stored on Amazon S3 Bucket.
2) Model Training
- Labeled data obtained from step 1 is used to train supervised machine learning models. Training happens on Amazon ECS clusters using a custom training algorithm. Training algorithms are dockerized and the image is stored in Amazon Elastic Container Registry (Amazon ECR).
- The trained parameters which are obtained from model training are stored in an S3 bucket.
3) Model deployment
- Model deployment and inference take place on Amazon SageMaker.
- To ensure consistent authentication with all the Polly services, we don’t expose SageMaker endpoints directly to the users. Instead, users can interact with these ML models through Polly APIs and SDKs.
Workstream 4 – Scaling computational workloads
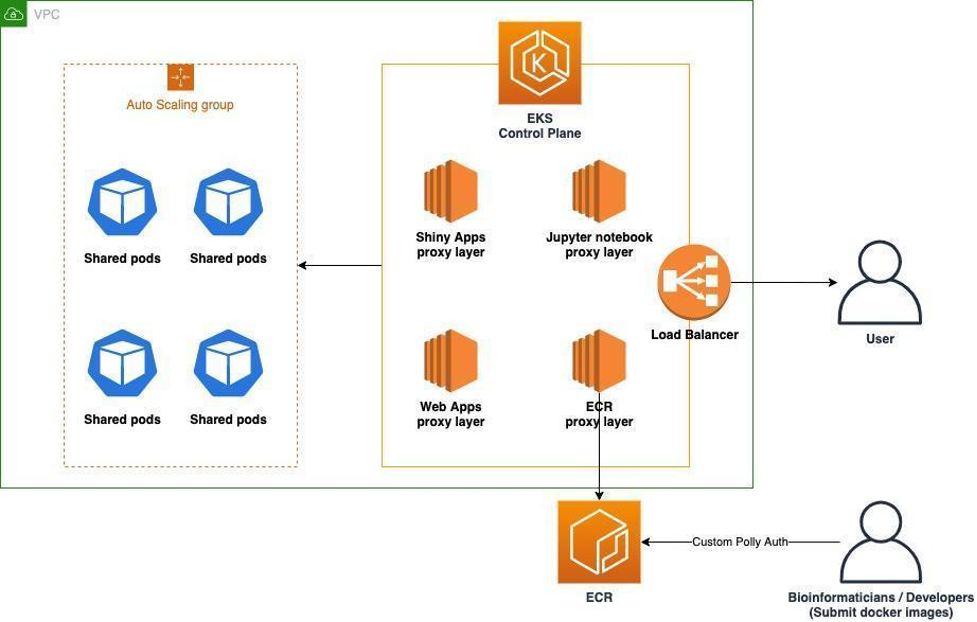
There are many ways our users consume data on Polly. It can be through web apps, R-based Shiny apps, command-line jobs, or Jupyter notebooks. Self-hosting these applications can become challenging as there are many aspects that need to be managed, like scalability, availability, deployments, and collaboration. Polly solves these challenges by providing a managed solution that does the undifferentiated heavy lifting for the user. For hosting applications, users can directly upload docker images on the platform. For using Ipython notebooks, they can create a new notebook using Polly UI. Additionally, users can choose the size of the machine for applications, jobs, and Jupyter notebooks.
For enabling scalable computational applications:
- Polly uses Amazon ECR to store applications and workflows in the form of dockerized images.
- Polly extensively uses Amazon Elastic Kubernetes Service (Amazon EKS) to deploy applications. The orchestration and scaling happen on Amazon EKS.
- Jupyter hub is integrated with EKS to deploy and scale Jupyter applications and there is a custom proxy layer that is used to scale the rest of the dockerized application on EKS which internally consumes EKS API.
- A combination of AWS Lambda and API gateway is used for the API interface.
- Users just have to upload a docker image to Amazon ECR using their Polly credentials. The shared instances/pods are used to allocate resources.
Elucidata’s goal has been to double the data on Polly every quarter. Hence, it’s pertinent to create data and machine learning pipelines needing minimal human intervention so that the increase in data does not necessitate an increase in team size. When the rate of ingestion can vary from zero bytes on a particular day to TBs on another day, it’s crucial to be able to scale seamlessly. With AWS as a partner, we have been able to utilize the elasticity of the cloud effectively; and adapt to whatever the data may throw at us.
Learn more about Amazon Athena, an interactive query service that makes it easy to analyze data in Amazon S3 using standard SQL. Athena is serverless, so there is no infrastructure to manage, and you pay only for the queries that you run.
 Prior to starting Elucidata, Swetabh Pathak was a founding member and product manager at start-ups in technology and affordable STEM education, and has an integrated Masters degree in Mathematics and Computer Science from IIT Delhi.
Prior to starting Elucidata, Swetabh Pathak was a founding member and product manager at start-ups in technology and affordable STEM education, and has an integrated Masters degree in Mathematics and Computer Science from IIT Delhi.
 Sahil Rai is an Engineering Manager at Elucidata. He leads the data infrastructure team managing critical data pipelines and storage infrastructure. He has extensive experience architecting and building solutions on AWS.
Sahil Rai is an Engineering Manager at Elucidata. He leads the data infrastructure team managing critical data pipelines and storage infrastructure. He has extensive experience architecting and building solutions on AWS.