AWS Architecture Blog
Genomics workflows, Part 1: automated launches
Genomics workflows are high-performance computing workloads. Traditionally, they run on-premises with a collection of scripts. Scientists run and manage these workflows manually, which slows down the product development lifecycle. Scientists spend time to administer workflows and handle errors on a day-to-day basis. They also lack sufficient compute capacity on-premises.
In Part 1 of this series, we demonstrate how life sciences companies can use Amazon Web Services (AWS) to remove the traditional heavy lifting associated with genomic studies. We use AWS Step Functions to orchestrate workflow steps, including error handling. With AWS Batch, we horizontally scale-out the analytic tasks for optimal performance. This allows genome scientists to focus on scientific discovery while AWS runs their workflows.
Use case
Workflow systems used for genomic analysis include Cromwell, Nextflow, and regenie. These high-performance computing systems share the following requirements:
- Fast access to datasets at petabyte scale
- Parallel task distribution, with horizontal compute scale-out
- Data processing in batches following a specific sequence of data analysis steps, which vary by use case
We explore the use case of regenie. regenie is a common, open-source utility for whole-genome regression modelling of large genome-wide association studies (GWAS). GWAS compare DNA datasets of individuals with a specific trait or disease. The intent is to associate the identified trait/disease with DNA variants. Among other positive results, this helps identify at-risk patients, plus testing and prevention opportunities.
regenie is a C++ program that runs in two steps:
- The first step searches for variants associated with a specific trait in a dataset of individuals with the trait, in order to create a whole-genome regression model that captures the variance.
- The second step validates for association with the identified variants against a larger dataset, typically in the scale of petabytes, and launches a sequence of tasks run on data batches.
Solution overview
The entire regenie workflow and associated tasks of attaching and deleting file-share access to sample data, as well as spinning up compute instances for parallel computing, can be orchestrated with Step Functions. We use Amazon FSx for Lustre as a high-performance, transient file system providing file access to the datasets stored in an Amazon Simple Storage Service (Amazon S3) bucket. AWS Batch allows us to programmatically spin up multiple Amazon Elastic Compute Cloud (Amazon EC2) instances on which regenie can distribute parallel computing tasks. We do this with an AWS Lambda function that calculates the number of required batch jobs based on the requested size of samples per batch.
regenie is available as Docker image on GitHub. We push the image to Amazon Elastic Container Registry from which AWS Batch can pull it with the creation of new jobs at launch time. The Step Functions state machine is initiated by a Lambda function, with interactive user input. In the past, scientists have also directly interacted with the Step Functions API via the AWS Management Console or by running start-execution in the AWS Command Line Interface and passing a JSON file with the input parameters.
Amazon CloudWatch provides a consolidated overview of performance metrics, including elapsed time, failed jobs, and error types. You can keep logs of your failed jobs in Amazon CloudWatch Logs (Figure 1). You can set up filters to match specific error types, plus create subscriptions to deliver a real-time stream of your log events to Amazon Kinesis or AWS Lambda for further retry.
Alternatively, the Step Functions workflow triggers another Lambda function, which puts failed job logs to Amazon DynamoDB. In the past, we have used this to ease data access and manipulation via the AWS management console. Scientists updated table items and DynamoDB Streams initiated the retry.
Workflow automation
With each invocation, Step Functions initiates a new instance of the state machine. AWS documentation provides an overview of the API quotas. Step Functions allows the modeling of the entire workflow, including custom application error handling. Map state improves performance by parallelizing workflow branches.
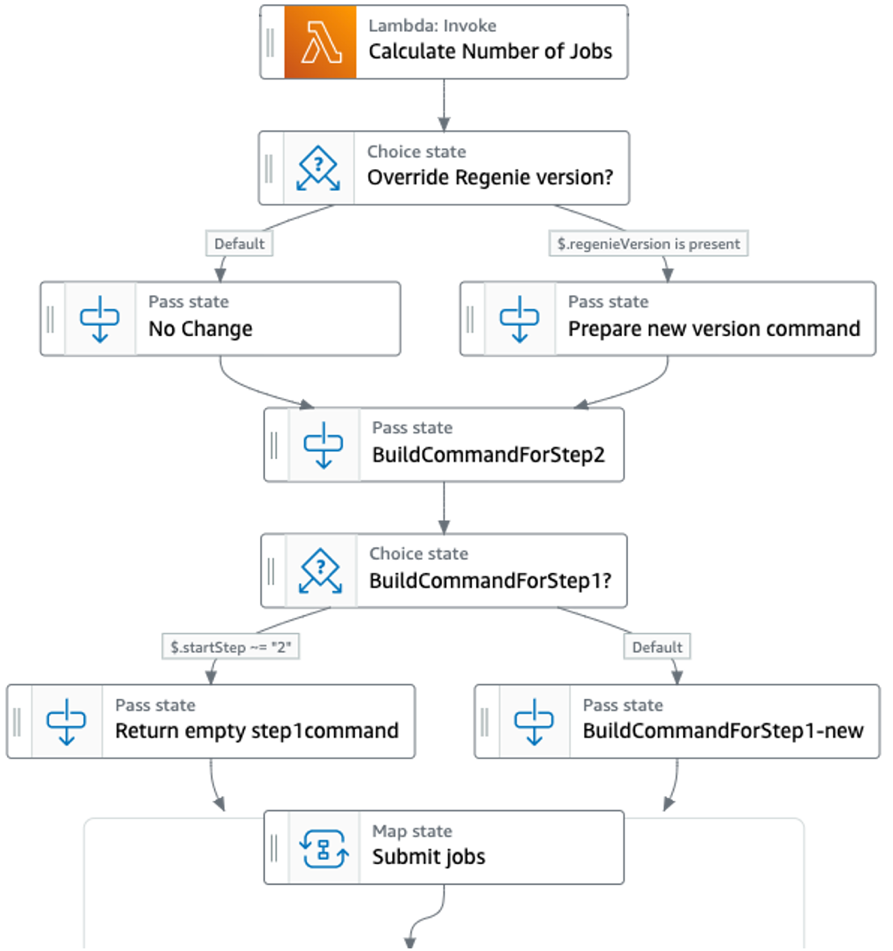
The state machine initiates the build of the file system and, once it’s ready, creates a data repository association with the sample data stored on Amazon S3. It waits until the data repository association is complete and proceeds with the calculation of batch jobs, based on a user-defined number of samples to be processed per batch job (Figure 2). This is essential to determine the amount of compute instances required for data processing.
Next, the state machine builds the commands to launch the regenie steps, as requested by the user, and submit the jobs for AWS Batch (Figure 3). The workflow checks if a specific version of regenie was requested by the user, otherwise, it defaults to the version of regenie on the container.
Then, we build the commands to initiate the two regenie steps. Step 2 may need to run in multiple iterations on different datasets (more often than Step 1). This is also determined with user input at initiation of the workflow. With Step Functions, we create runner logic to build the set of commands dynamically. This pattern is applicable to other scientific workloads, as well.
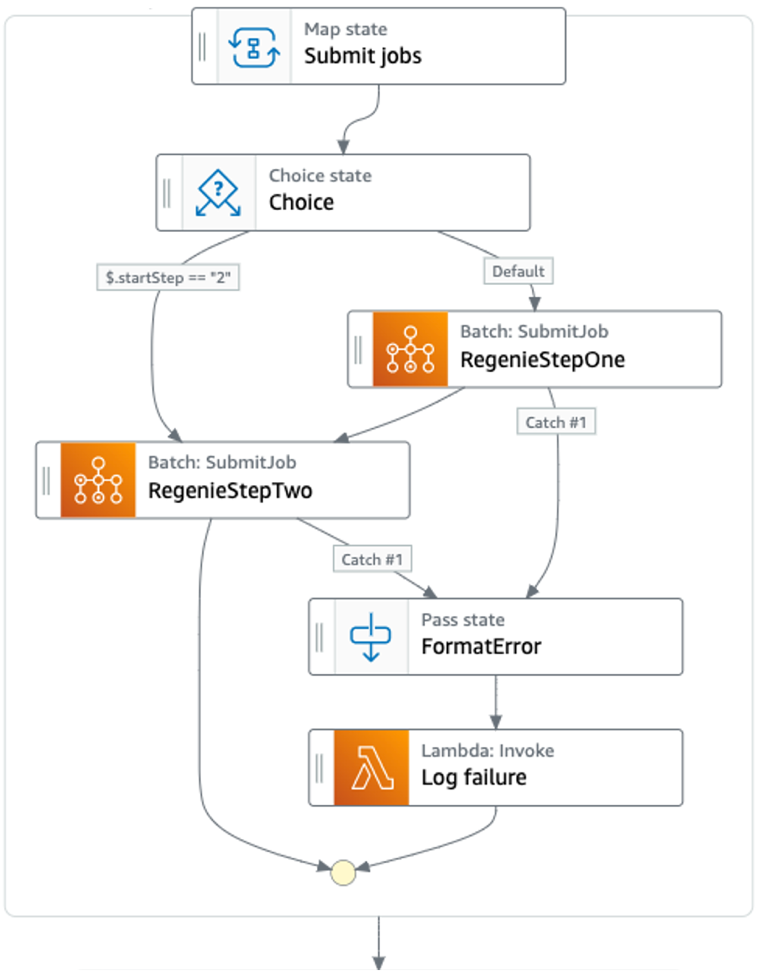
Once jobs are submitted, the workflow proceeds (by default) with the initiation of Step 1 of regenie; if requested by the user, the workflow will proceed directly to step 2 (Figure 4).
Any errors during batch launch leading to the failure of a job are passed, in this case, to a Lambda function. We configure the Lambda function to write the failed job logs to Amazon DynamoDB or as S3 objects.
Finally, the Step Functions workflow checks for pending errors and confirms that all jobs have finished their initiation. Then, it deletes the file system and data repository association and ends the workflow instance (Figure 5).
As demonstrated, we can automate the entire process, from data access to verifying job completion and cleaning-up transient resources. This removes manual error handling and retry, plus reduces the overall cost of running regenie workflows. We also showed in Figure 3 that you can build commands dynamically for different scientific workloads.
Conclusion
In this blog post, we addressed a common pain point in the daily work of life sciences research teams. Traditionally, they had to run genomics workflows manually on limited compute capacity. Moving those workflows to AWS eliminates the heavy lifting of running scripts manually and expedites computational cycles. This allows research teams to stay focused on scientific discovery.
We recommend a thorough performance testing when setting up your genomics workflows. This includes determining the most suitable EC2 instance size. Some workflows, such as regenie, are single-threaded and benefit from horizontal scale-out of the number of instances but not from vertical scale-out of instance sizes.
Related information
- Genomics workflows, Part 2: simplify Snakemake launches
- Genomics workflows, Part 3: automated workflow manager
- Genomics workflows, Part 4: processing archival data
- Genomics workflows, Part 5: automated benchmarking
- Genomics workflows, Part 6: cost prediction
- Genomics workflows, Part 7: analyze public RNA sequencing data using AWS HealthOmics
- Genomics on AWS
- Amazon Genomics CLI