AWS Big Data Blog
Run Common Data Science Packages on Anaconda and Oozie with Amazon EMR
In the world of data science, users must often sacrifice cluster set-up time to allow for complex usability scenarios. Amazon EMR allows data scientists to spin up complex cluster configurations easily, and to be up and running with complex queries in a matter of minutes.
Data scientists often use scheduling applications such as Oozie to run jobs overnight. However, Oozie can be difficult to configure when you are trying to use popular Python packages (such as “pandas,” “numpy,” and “statsmodels”), which are not included by default.
One such popular platform that contains these types of packages (and more) is Anaconda. This post focuses on setting up an Anaconda platform on EMR, with an intent to use its packages with Oozie. I describe how to run jobs using a popular open source scheduler like Oozie.
Walkthrough
For this post, you walk through the following tasks:
- Create an EMR cluster.
- Download Anaconda on your master node.
- Configure Oozie.
- Test the steps.
Create an EMR cluster
Spin up an Amazon EMR cluster using the console or the AWS CLI. Use the latest release, and include Apache Hadoop, Apache Spark, Apache Hive, and Oozie.
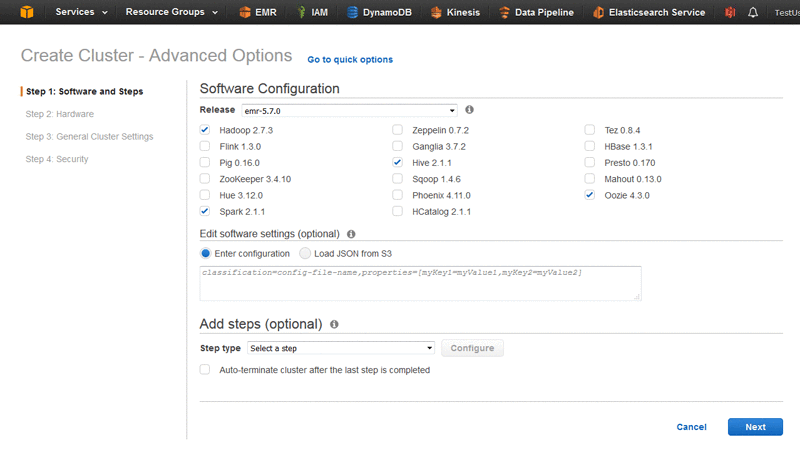
To create a three-node cluster in the us-east-1 region, issue an AWS CLI command such as the following. This command must be typed as one line, as shown below. It is shown here separated for readability purposes only.
One-line version for reference:
Download Anaconda
SSH into your EMR master node instance and download the official Anaconda installer:
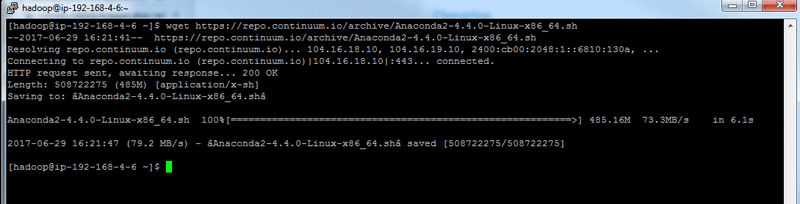
At the time of publication, Anaconda 4.4 is the most current version available. For the download link location for the latest Python 2.7 version (Python 3.6 may encounter issues), see https://www.continuum.io/downloads. Open the context (right-click) menu for the Python 2.7 download link, choose Copy Link Location, and use this value in the previous wget command.
This post used the Anaconda 4.4 installation. If you have a later version, it is shown in the downloaded filename: “anaconda2-<version number>-Linux-x86_64.sh”.
Run this downloaded script and follow the on-screen installer prompts.
For an installation directory, select somewhere with enough space on your cluster, such as “/mnt/anaconda/”.
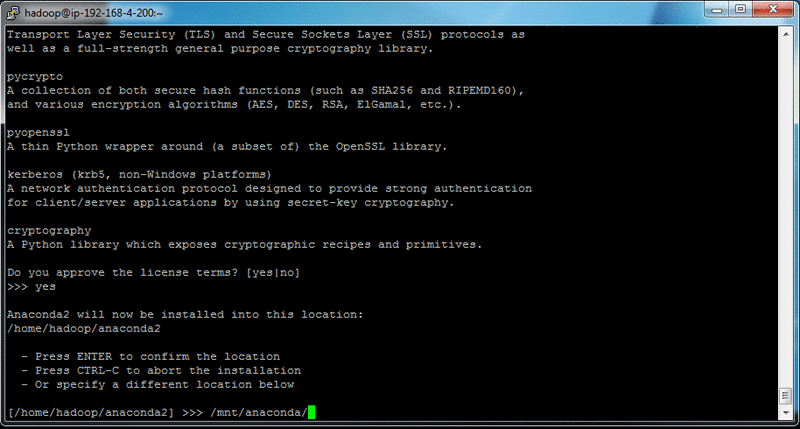
The process should take approximately 1–2 minutes to install. When prompted if you “wish the installer to prepend the Anaconda2 install location”, select the default option of [no].
After you are done, export the PATH to include this new Anaconda installation:
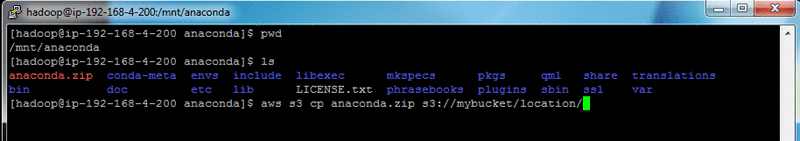
Zip up the Anaconda installation:
The zip process may take 4–5 minutes to complete.
(Optional) Upload this anaconda.zip file to your S3 bucket for easier inclusion into future EMR clusters. This removes the need to repeat the previous steps for future EMR clusters.
Configure Oozie
Next, you configure Oozie to use Pyspark and the Anaconda platform.
Get the location of your Oozie sharelibupdate folder. Issue the following command and take note of the “sharelibDirNew” value:
For this post, this value is “hdfs://ip-192-168-4-200.us-east-1.compute.internal:8020/user/oozie/share/lib/lib_20170616133136”.

Pass in the required Pyspark files into Oozies sharelibupdate location. The following files are required for Oozie to be able to run Pyspark commands:
- pyspark.zip
- py4j-0.10.4-src.zip
These are located on the EMR master instance in the location “/usr/lib/spark/python/lib/”, and must be put into the Oozie sharelib spark directory. This location is the value of the sharelibDirNew parameter value (shown above) with “/spark/” appended, that is, “hdfs://ip-192-168-4-200.us-east-1.compute.internal:8020/user/oozie/share/lib/lib_20170616133136/spark/”.
To do this, issue the following commands:
After you’re done, Oozie can use Pyspark in its processes.
Pass the anaconda.zip file into HDFS as follows:
(Optional) Verify that it was transferred successfully with the following command:
On your master node, execute the following command:
Set the PYSPARK_PYTHON environment variable on the executor nodes. Put the following configurations in your “spark-opts” values in your Oozie workflow.xml file:
This is referenced from the Oozie job in the following line in your workflow.xml file, also included as part of your “spark-opts”:
Your Oozie workflow.xml file should now look something like the following:
Test steps
To test this out, you can use the following job.properties and myPysparkProgram.py file, along with the following steps:
job.properties
Note: You can get your master node IP address (denoted as “ip-xxx-xxx-xxx-xxx” here) from the value for the sharelibDirNew parameter noted earlier.
myPysparkProgram.py
Put the “myPysparkProgram.py” into the location mentioned between the “<jar>xxxxx</jar>” tags in your workflow.xml. In this example, the location is “hdfs:///user/oozie/apps/”. Use the following command to move the “myPysparkProgram.py” file to the correct location:
Put the above workflow.xml file into the “/user/oozie/apps/” location in hdfs:
Note: The job.properties file is run locally from the EMR master node.
Create a sample input.txt file with some data in it. For example:
input.txt
Put this file into hdfs:
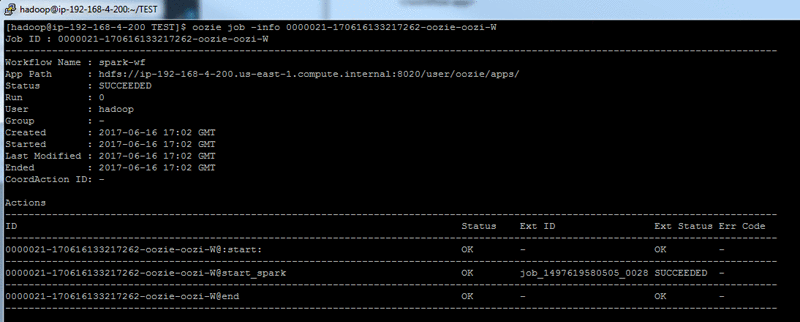
Execute the job in Oozie with the following command. This creates an Oozie job ID.

You can check the Oozie job state with the command:
- When the job is successfully finished, the results are located at:
- Run the following commands to view the output:
The output will be:
Summary
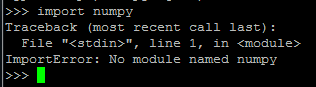
The myPysparkProgram.py has successfully imported the numpy library from the Anaconda platform and has produced some output with it. If you tried to run this using standard Python, you’d encounter the following error:
Now when your Python job runs in Oozie, any imported packages that are implicitly imported by your Pyspark script are imported into your job within Oozie directly from the Anaconda platform. Simple!
If you have questions or suggestions, please leave a comment below.
Additional Reading
Learn how to use Apache Oozie workflows to automate Apache Spark jobs on Amazon EMR.

About the Author
 John Ohle is an AWS BigData Cloud Support Engineer II for the BigData team in Dublin. He works to provide advice and solutions to our customers on their Big Data projects and workflows on AWS. In his spare time, he likes to play music, learn, develop tools and write documentation to further help others – both colleagues and customers alike.
John Ohle is an AWS BigData Cloud Support Engineer II for the BigData team in Dublin. He works to provide advice and solutions to our customers on their Big Data projects and workflows on AWS. In his spare time, he likes to play music, learn, develop tools and write documentation to further help others – both colleagues and customers alike.