AWS HPC Blog
Tag: GROMACS
Large scale, cost effective GROMACS simulations using the Cyclone Solution from AWS
Proteins hold the secrets of life, if only we can study them in sufficient detail. Learn how a team at Max Planck leveraged AWS Spot Instances with Cyclone to run massive GROMACS simulations and push the boundaries of biophysics.
Best practices for running molecular dynamics simulations on AWS Graviton3E
If you run molecular dynamics simulations, you need to read this. We walk through running benchmarks of popular apps like GROMACS and LAMMPS on new Hpc7g instances and Graviton3E processors. The results – up to 35% better vector performance versus Graviton3! Learn how to optimize your own workflows.
Instance sizes in the Amazon EC2 Hpc7 family – a different experience
Hpc7g is the first Amazon EC2 HPC instance offering with multiple instance sizes, but this is quite different from the experience of getting smaller instances from other non-HPC instance families. Today, we want to take a moment to explore why this is different, and how it helps.
Running cost-effective GROMACS simulations using Amazon EC2 Spot Instances with AWS ParallelCluster
In this blog post, we cover how to run GROMACS – a popular open source designed for simulations of proteins, lipids, and nucleic acids – cost effectively by leveraging EC2 Spot Instances within AWS ParallelCluster. We also show how to checkpoint GROMACS to recover gracefully from possible Spot Instance interruptions.
GROMACS performance on Amazon EC2 with Intel Ice Lake processors
We recently launched two new Amazon EC2 instance families based on Intel’s Ice Lake – the C6i and M6i. These instances provide higher core counts and take advantage of generational performance improvements on Intel’s Xeon scalable processor family architectures. In this post we show how GROMACS performs on these new instance families. We use similar methodologies as for previous posts where we characterized price-performance for CPU-only and GPU instances (Part 1, Part 2, Part 3), providing instance recommendations for different workload sizes.
Running 20k simulations in 3 days to accelerate early stage drug discovery with AWS Batch
In this blog post, we’ll describe an ensemble run of 20K simulations to accelerate the drug discovery process, while also optimizing for run time and cost. We used two popular open-source packages — GROMACS, which does a molecular dynamics simulations, and pmx, a free-energy calculation package from the Computational Biomolecular Dynamics Group at Max Planck Institute in Germany.
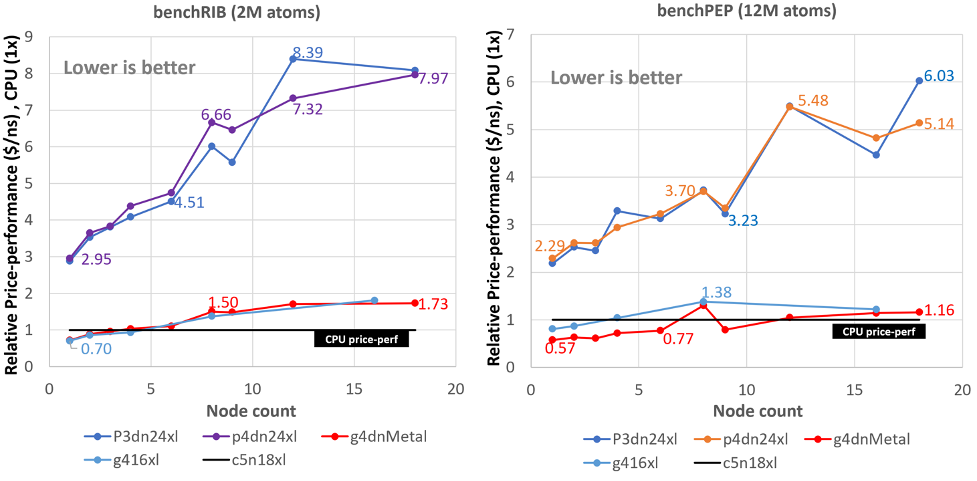
Running GROMACS on GPU instances: multi-node price-performance
This three-part series of posts cover the price performance characteristics of running GROMACS on Amazon Elastic Compute Cloud (Amazon EC2) GPU instances. Part 1 covered some background no GROMACS and how it utilizes GPUs for acceleration. Part 2 covered the price performance of GROMACS on a particular GPU instance family running on a single instance. […]
Running GROMACS on GPU instances: single-node price-performance
This three-part series of posts cover the price performance characteristics of running GROMACS on Amazon Elastic Compute Cloud (Amazon EC2) GPU instances. Part 1 covered some background no GROMACS and how it utilizes GPUs for acceleration. This post (Part 2) covers the price performance of GROMACS on a particular GPU instance family running on a […]
Running GROMACS on GPU instances
Comparing the performance of real applications across different Amazon Elastic Compute Cloud (Amazon EC2) instance types is the best way we’ve found for finding optimal configurations for HPC applications here at AWS. Previously, we wrote about price-performance optimizations for GROMACS that showed how the GROMACS molecular dynamics simulation runs on single instances, and how it […]
GROMACS price-performance optimizations on AWS
Molecular dynamics (MD) is a simulation method for analyzing the movement and tracing trajectories of atoms and molecules where the dynamics of a system evolve over time. MD simulations are used across various domains such as material sciences, biochemistry, biophysics and are typically used in two broad ways to study a system. The importance of […]