AWS Compute Blog
Folding@home infectious disease research with Spot Instances
This post was contributed by Jarman Hauser, Jessie Xie, and Kinnar Kumar Sen.
Folding@home (FAH) is a distributed computing project that uses computational modeling to simulate protein structure, stability, and shape (how it folds). These simulations help to advance drug discoveries and cures for diseases linked to protein dynamics within human cells. The FAH software crowdsources its distributed compute platform allowing anyone to contribute by donating unused computational resources from personal computers, laptops, and cloud servers.
Overview
In this post, I walk through deploying EC2 Spot Instances, optimized for the latest Folding@home client software. I describe how to be flexible across a combination of GPU-optimized Amazon EC2 Spot Instances configured in an EC2 Auto Scaling group. The Auto Scaling group handles launching and maintaining a desired capacity, and automatically request resources to replace any that are interrupted or manually shut down.
Spot Instances are spare EC2 capacity available at up to a 90% discount compared to On-Demand Instance prices. The only difference between On-Demand Instance and Spot Instances is that Spot Instances can be interrupted by EC2 with two minutes of notification when EC2 needs the capacity back. This makes Spot Instances a great fit for stateless, fault-tolerant workloads like big data, containers, batch processing, AI/ML training, CI/CD and test/dev. For more information, see Amazon EC2 Spot Instances.
In addition to being flexible across instance types, another best practice for using Spot Instances effectively is to select the appropriate allocation strategy. Allocation strategies in EC2 Auto Scaling help you automatically provision capacity according to your workload requirements. We recommend that using the capacity optimized strategy to automatically provision instances from the most-available Spot Instance pools by looking at real-time capacity data. Because your Spot Instance capacity is sourced from pools with optimal capacity, this decreases the possibility that your Spot Instances are reclaimed. For more information about allocation strategies, see Spot Instances in the EC2 Auto Scaling user guide and configuring Spot capacity optimization in this user guide.
What you’ll build
- An Amazon Virtual Private Cloud (VPC) configured with public and private subnets according to AWS best practices.
- Identity and Access Management (IAM) roles to manage permissions for EC2 Auto Scaling.
- Security group for the EC2 Spot Instancesto control inbound and outbound traffic
- Auto Scaling group for scaling EC2 Spot Instancesin and out as needed using the capacity-optimized allocation strategy.
Amazon CloudWatch instance metrics and logs for real-time monitoring of the protein folding progress.
To complete the setup, you must have an AWS account with permissions to the listed resources above. When you sign up for AWS, your AWS account is automatically signed up for all services in AWS, including Amazon EC2. If you don’t have an AWS account, find more info about creating an account here.
Costs and licensing
The AWS CloudFormation (CFn) template includes customizable configuration parameters. Some of these settings, such as instance type, affect the cost of deployment. For cost estimates, see the pricing pages for each AWS service you are using. Prices are subject to change. You are responsible for the cost of the AWS services used. There is no additional cost for using the CFn template.
Note: There is no additional charge to use Deep Learning AMIs — you pay only for the AWS resources while they’re running. Folding@home client software is a free, open-source software that is distributed under the Folding@home EULA.
Tip: After you deploy the AWS CloudFormation template, we recommend that you enable AWS Cost Explorer. Cost Explorer is an easy-to-use interface that lets you visualize, understand, and manage your AWS costs and usage e.g. you can break down costs to show hourly costs for your protein folding project.
How to deploy
Part one: Download and configure the CFn template
First thing you must do is download, then make a few edits to the template.
Once downloaded, open the template file in your favorite text editor to make a few edits to the configuration before deploying.
In the User Information section, you have the option to create a unique user name, join or create a new team, or contribute anonymously. For this example, I leave the values set to default and contribute as an anonymous user, the default team. More details about teams and leaderboards can be found here and details about PASSKEYs here.
Once edited and saved to a location you can easily find later, in the next section you’ll learn how to upload the template in the AWS CloudFormation console.
Part two: Launching the stack
Next, log into the AWS Management Console, choose the Region you want to run the solution in, then navigate to AWS CloudFormation to launch the template.
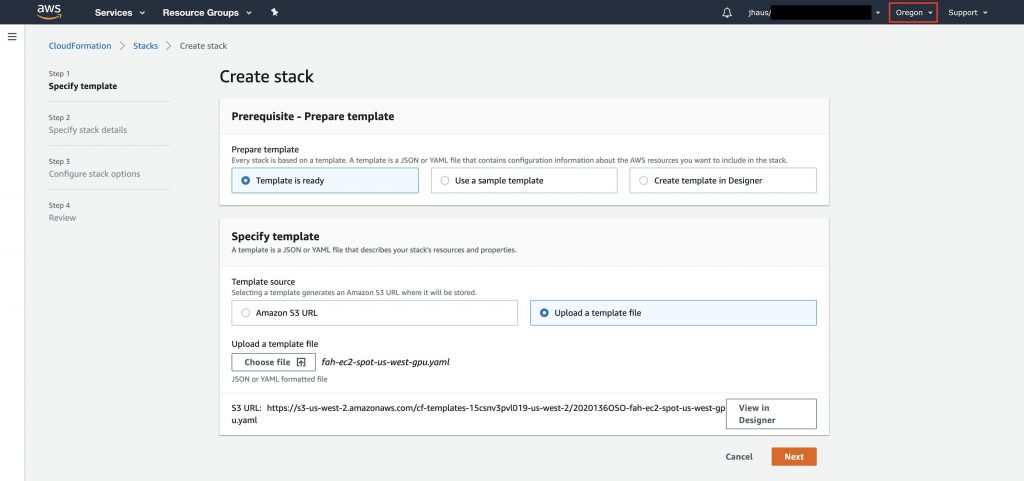
In the AWS CloudFormation console, click on Create stack. Upload the template we just configured and click on Next to specify stack details.
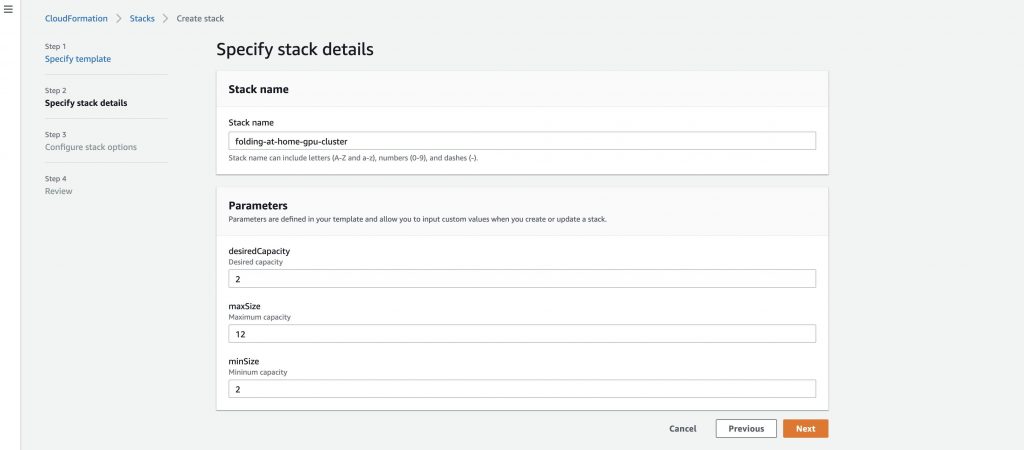
Enter a stack name and adjust the capacity parameters as needed. In this example I set the desiredCapacity and minSize at 2 to handle protein folding jobs assigned to the client, and then the maxSize set at 12. Setting your maxSize to 12 ensures you have capacity for larger jobs that get assigned. These parameters can be adjusted based on your desired capacity.
If breaking out usage and cost data is required,, you can optionally add additional configurations like tags, permissions, stack policies, rollback options, and more advanced options in the next stack configuration step. Click Next to Review and then create the stack.
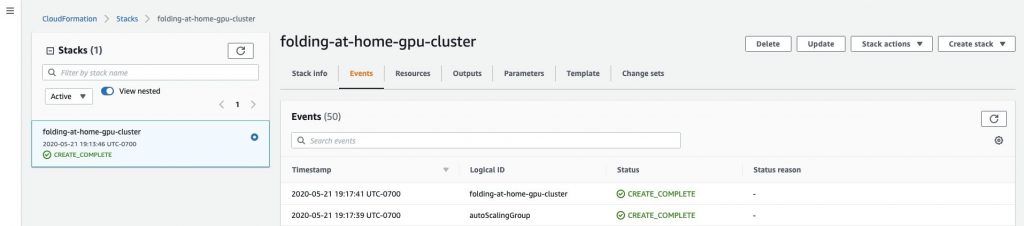
Under the Events tab, you can see the status of the AWS resources being created. When the status is CREATE_COMPLETE (approx. 3–5 minutes), the environment with Folding@home is installed and ready. Once the stack is created, the GPU instances will begin protein simulation.
Results
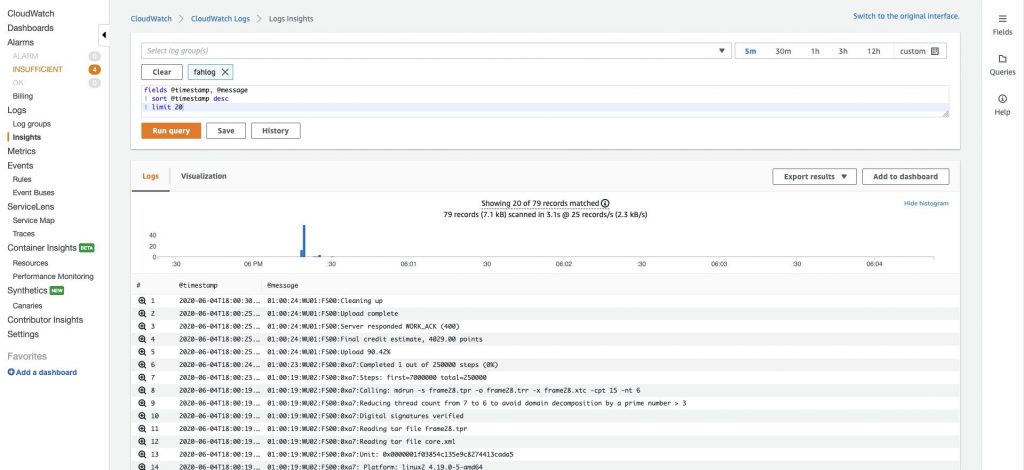
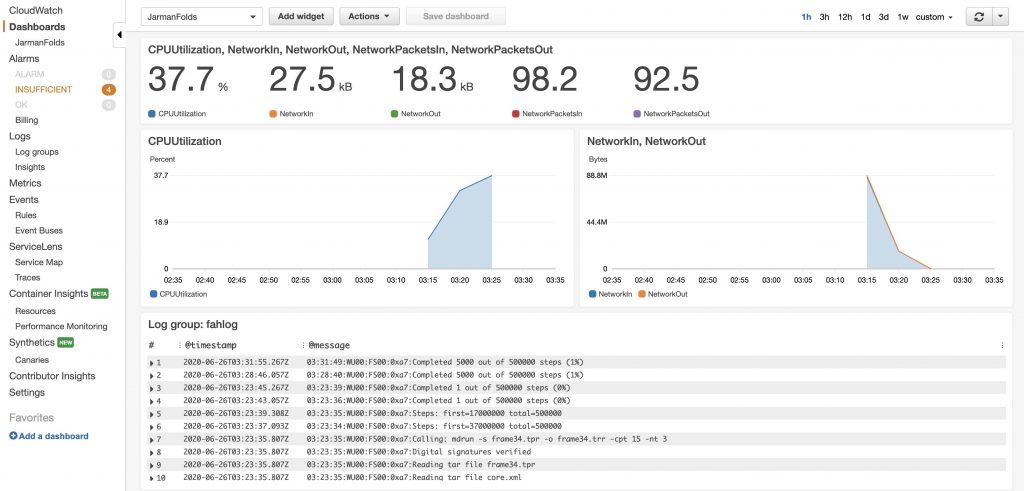
The AWS CloudFormation template creates a log group “fahlog” that each of the instances send log data to. This allows you to visualize the protein folding progress in near real time via the Amazon CloudWatch console. To see the log data, navigate over to the Resources tab and click on the cloudWatchLogGroup link for ‘fahlog’. Alternatively, you can navigate to the Amazon CloudWatch console and choose ‘fahlog’ under log groups. Note: Sometimes it takes a bit of time for Folding@Home Work Units (WU) to be downloaded in the instances and allocate all the available GPUs.
In the CloudWatch console, check out the Insights feature in the left navigation menu to see analytics for your protein folding logs. Select ‘fahlog’ in the search box and run the default query that is provided for you in the query editor window to see your protein folding results.
Another thing you can do is create a dashboard in the CloudWatch console to automatically refresh based on the time intervals you set. Under Dashboards in the left navigation bar, I was able to quickly create a few widgets to visualize CPU utilization, network in/out, and protein folding completed steps. This is a nifty tool that, with a little more time, you could configure more detailed metrics like cost per fold, and GPU monitoring.
Part three: Clean up
You can let this run as long as you want to contribute to this project. When you’re ready to stop, AWS CloudFormation gives us the option to delete the stack and resources created. On the AWS CloudFormation console, select the stack, and select delete. When you delete a stack, you delete the stack and all of its resources.
Conclusion
In this post, I shared how to launch a cluster of EC2 GPU-optimized Spot Instances to aid in Folding@home’s protein dynamics research that could lead to therapeutics for infectious diseases. I leveraged Spot best practices by being flexible with instance selections across multiple families, sizes, and Availability Zones, and by choosing the capacity-optimized allocation strategy to ensure our cluster scales optimally and securely. Now you are ready to donate compute capacity with Spot Instances to aid disease research efforts on Folding@home.
About Folding@home
Folding@home is currently based at the Washington University School of Medicine in St. Louis, under the directorship of Dr. Greg Bowman. The project was started by the Pande Laboratory at Stanford University, under the direction of Dr. Vijay Pande, who led the project until 2019.[4] Since 2019, Folding@home has been led by Dr. Greg Bowman of Washington University in St. Louis, a former student of Dr. Pande, in close collaboration with Dr. John Chodera of MSKCC and Vince Voelz of Temple University.[5]
With heightened interest in the project, Folding@home has grown to a community of 2M+ users, bringing together the compute power of over 600K GPUs and 1.6M CPUs.
This outpouring of support has made Folding@home one of the world’s fastest computing systems – achieving speeds of approximately 1.2 exaFLOPS, or 2.3 x86 exaFLOPS, by April 9, 2020 – making it the world’s first exaFLOP computing system. Folding@home‘s COVID-19 effort specifically focuses on better understanding how the viral protein’s moving parts enable to infect a human host, evade an immune response, and create new copies of the virus. The project is leveraging this insight to help design new therapeutic antibodies and small molecules that might prevent infection. They are engaged with a number of experimental collaborators to quickly iterate between computational design and experimental testing.