AWS Open Source Blog
How DNAnexus used the open source Former2 project to create infrastructure as code templates for their disaster recovery pipeline
This is a guest post by Varderes Barsegyan, a former DevOps engineer at DNAnexus.
Founded in 2009, APN Partner DNAnexus is a cloud-based genomics company that has built a cloud platform and global network for scientific collaboration and accelerated discovery. The platform provides compute and storage capabilities, customizable bioinformatics pipelines, and supports customers with specialized needs via the xVantage solution science group. Born in the cloud, DNAnexus has been running on AWS since 2009. The critical pieces of the platform were captured using the infrastructure as code design. However, other portions of the environment would be more time consuming to rebuild in the event of a disaster. Driven by our FedRAMP certification and desire to improve the reliability of the platform continuously, we decided to create a solution. We would continuously back up of all of our AWS resources as Terraform scripts.
With the help of our AWS account team, we began our search by exploring two leading enterprise options for cloud backup and disaster recovery (DR). Though comprehensive in their capabilities, that these solutions would not be a good fit for DNAnexus became clear, due to the following reasons:
- They would not easily integrate with the current deployment and maintenance pipelines because of our custom configuration management and infrastructure tooling software.
- They offered baseline capabilities that either did not apply to our use case or went far beyond our needs.
The search then turned to open source tools, leading us to evaluate Former2 as a potential solution. Former2 is an open source DR tool created by Ian McKay that generates AWS CloudFormation or Terraform templates from existing AWS resources. We performed an extensive evaluation of Former2 by first capturing resources in our environment and storing them in Terraform configuration files. We then deleted these resources from AWS and restored them with Terraform. Furthermore, we learned that Former2 would easily integrate with our platform and pipelines for a few reasons:
- The DNAnexus ops and engineering teams already use Terraform extensively.
- DNAnexus did not need to back up the resources themselves, but rather the configuration of those resources.
- Due to the urgency of this project, we needed a solution that could be deployed and maintained quickly and easily.
- As a result of our highly specialized configuration management and infrastructure tooling software, we needed a solution that could be integrated into our pipelines easily.
This blog post outlines the solution that we created using Former2 to regularly capture and archive DNAnexus platform resources and configurations. We explain how Former2 was integrated with our platform, what challenges we faced during the integration process, and the future work we will perform to enhance our solution.
Solution overview
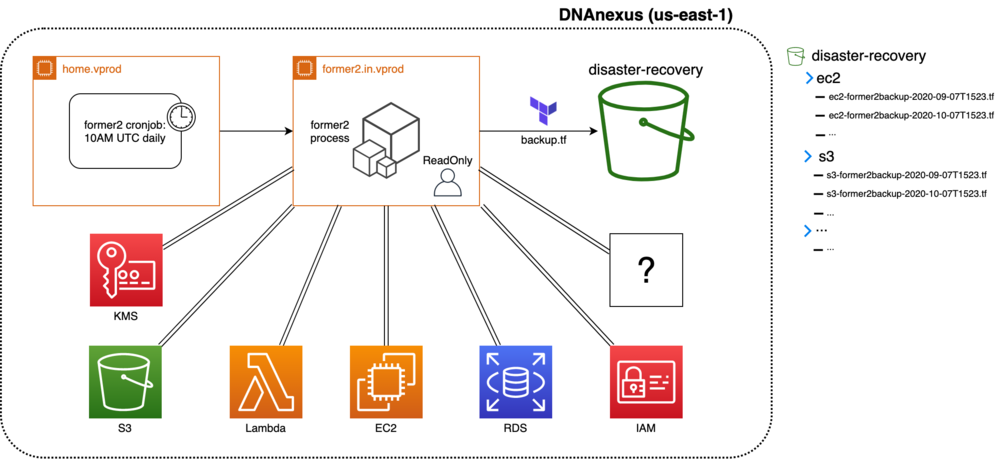
The architecture diagram displays the various components of the solution. We use and deploy Former2 in our environments through the following:
- We provision, configure, and stop an Ubuntu 18.04-based c5.4xlarge instance. We chose this instance due to the highly parallel nature of Former2 and the large number of AWS resources currently deployed in our environments.
- Once a day, a cron job launches this instance and deploys the Former2 jobs, one for each service type—Amazon Elastic Compute Cloud (Amazon EC2), VPC, Amazon Simple Storage Service (Amazon S3), etc.
- Each job generates its own Terraform configuration file containing all of the resources of that service.
- All of the resulting configuration files are copied into a dedicated DR bucket. Each service has its own directory in that bucket, and each backup file is named based on its date and time of creation.
- To complement these backups, we have written an extensive, easy-to-follow internal operations document that explains how to extract these templates and restore resources during emergency scenarios.
Implementation steps
Prerequisites
- Provision an AWS instance and install Node.js 12+, npm 6+, and Git. At DNAnexus, the dedicated Former2 instance is a c5.4xlarge running Ubuntu 18.04.
- Create a bucket (for example, DR).
- Create a read-only user (for example, former2) along with an API Access Key/Secret. To simplify, you can attach the AWS managed ReadOnlyAccess policy to this user. In case you would like to use a custom policy, make sure that the user has full read access to the resources of interest. No write access is required.
- Install the CLI version of Former2 on the instance:
Note: You can also deploy Former2 as a Docker container. See the Former2 CLI docs for more information.
- Finally, confirm that the security group of the instance running Former2 allows SSH access from the following instance.
Deployment
- On another instance with the AWS CLI installed, create a cron job similar to the following. In this example, the cron job runs daily at 10AM UTC:
- Create a script similar to the following that launches the instances and triggers the Former2 jobs:
- Create a script similar to the following, which runs on the instance via former2generate.sh script in step 2:
Implementation challenges
There were a number of challenges to work through while implementing this solution. Initially, Former2 did not scale to the size and scope of the AWS environment resources supporting the DNAnexus platform. When trying to capture all resources, Former2 crashed due to the size of the output file. Additionally, AWS API calls frequently failed due to too many simultaneous requests. Also there were features related to usability that were required for the solution to fit our needs. To support our use case, we made the following changes to the Former2 code base:
- We worked with the creator of Former2 to implement a fix to resolve the file creation issue for exceptionally large environments. In particular, we implemented a feature that enables you to exclude particular services such as CloudWatch. These services were unnecessary for us to capture, and they took up significant portions of the output.
- We then worked with the creator to implement a fix for the request limit exceptions that would cause failed requests to sleep and try again later.
- Due to the highly asynchronous nature of Former2, we customized the code base to set a random sleep between a specified time range during each AWS API call. Former2 was now able to handle RequestLimitExceeded exceptions and continuously retry failed API calls until they succeeded. However, we wanted to be good citizens and avoid this exception entirely. This minimized the risk of potential throttling and creating a bottleneck in another platform component.
- We requested and contributed to upstream changes in the Former2 CLI code based. These changes provided users with the ability to exclude and include services based on command-line arguments and filter IAM resources based on tags.
Conclusion
Harvesting and archiving the resources and configurations of the vast DNAnexus platform environment was a challenge. Fortunately, we were able to use Former2 to create Terraform templates representing our environment on a regular basis and archive them to support our DR efforts. Former2 provided us a highly customizable solution that we were able to integrate into our existing infrastructure. We were able to contribute features developed during this project back to the community.
With this solution in place, we have better visibility into the state of our environments and can directly access frequently updated configurations to deploy or restore resources. Furthermore, we can rely on Former2 to generate Terraform configurations reliably even for manually deployed resources. As a result, we have decreased our recovery time objective (RTO), a parameter that is critical to the success of our continuously growing platform.
Although this solution is a significant step in improving our DR processes, there is additional work to do to harness the power of these regular backups. For now, an on-call engineer must extract a backup from Amazon S3 and run Terraform manually for recovery. In the future, automation will either fully restore the appropriate resource, or notify our team that resources have been shut down. The automation will point you to the exact template needed to recover your resources. We will also put more validation in place for backups. These validations will ensure that each run captures the appropriate number of resources, and that our team receives notification on any incomplete backups caused by transient issues.
We also have begun working on a solution that uses serverless technologies, rather than having a Dedicated Instance set up to run Former2. We have modified the published Dockerfile for the containerized version of Former2 to write results to Amazon S3, and have set this up as a Fargate scheduled task. We will test this configuration and update this blog with code and instructions once this effort is completed.
The content and opinions in this post are those of the third-party author and AWS is not responsible for the content or accuracy of this post.
