AWS Public Sector Blog
Exatype: Cloud for HIV Drug Resistance Testing
Professor Simon Travers and his team of researchers based at the South African National Bioinformatics Institute (SANBI) at the University of the Western Cape (UWC) have launched the Exatype platform, which runs on AWS. The Exatype platform provides rapid, accurate HIV drug resistance analysis at affordable rates. Users upload HIV sequence data files directly from high-throughput sequencing machines and receive a detailed drug resistance genotype report for each sample.
What started as an idea from Professor Travers to help the over six million South Africans living with HIV has now gone commercial and can test hundreds of people per machine per day in a more cost effective, scalable way.
In 2011, when this idea became a reality, the team realized the number of approaches available for HIV drug resistance testing. They could use software with the sequencing machine, and then analyze the data locally. With this approach, they recognized support issues and the hurdle of keeping the software up to date, thus increasing the cost per test. Or they could perform the data analysis in the cloud. Without needing to buy servers, Professor Travers realized they could drive down the cost of data analysis substantially and have unlimited scalability.
“Being able to access the computation requirements we need for a specific amount of time instead of buying high-end computing hardware that we don’t use most of the time was a no brainer,” Professor Travers said. “When we migrated to AWS, we got the computing power we needed at a much cheaper cost.”
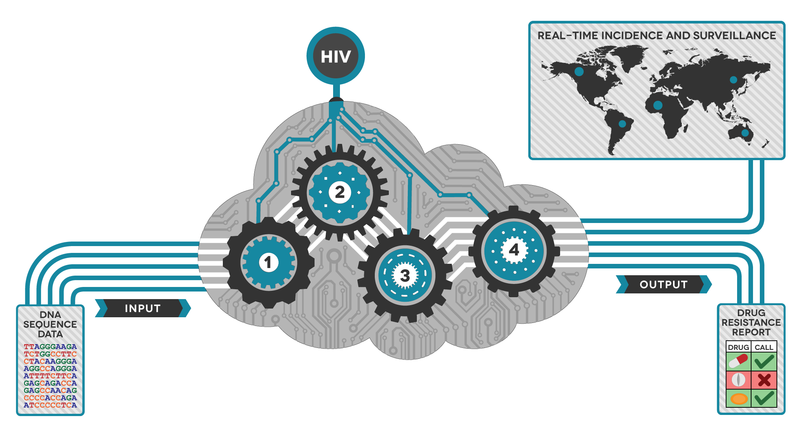
How does it work?
The goal of HIV drug resistance testing is to identify mutations present within the viral population of an HIV-infected individual. These mutations can cause resistance to one or more drugs, so accurately identifying their presence/absence can help determine the best path for treatment. Now with new ways of generating the sequencing data quickly, vast quantities of complex data is produced. Data production is relatively easy, but analyzing it can be difficult.
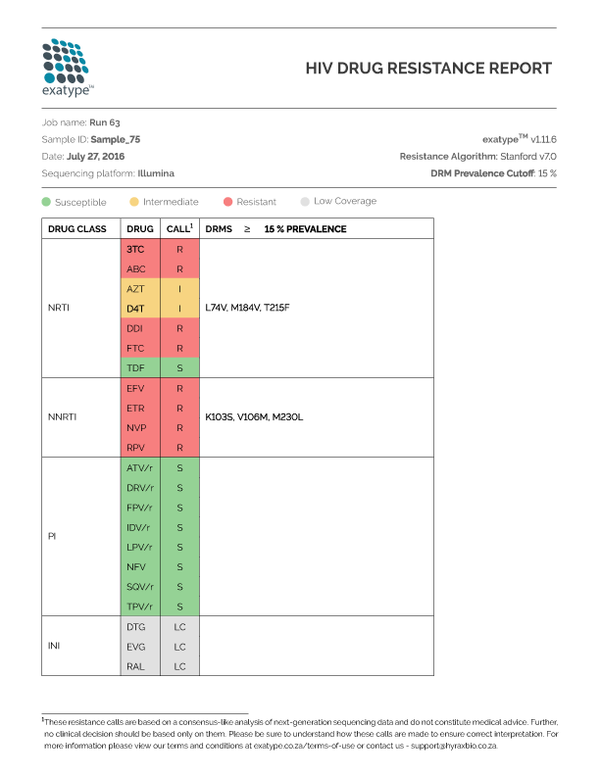
Simon and team have developed a tool that takes data directly off of the sequencing machine and produces an easy-to-read PDF report within an hour. But it isn’t just about the speed to result; Exatype is highly accurate and can distinguish between background errors in the data and true mutations.
“With AWS, we could analyze data from every HIV-positive person in the world and use every sequencing machine possible, and still be able to generate the reports without any delay,” Professor Travers said. “This is not possible with anything other than the cloud.”
In addition to being scalable and more cost effective, the current approach provides greater sensitivity to detect resistance at low prevalence. The traditional way identifies a drug resistance mutation only if it is present in greater than 20% of the virus in the individual. Theoretically, this new approach to sequencing means that drug-resistant viruses present in less than 1% of the viruses circulating within a HIV-infected individual can be identified.
Looking ahead – What’s next?
Based on the possibilities of the cloud, the team is looking to move into other microbial resistance disease lines, such as Staphylococcus aureus, tuberculosis, and Hepatitis C. Exatype is a disease-agnostic application, and the set of references and rules applied to various diseases can be altered without having to tailor computing requirements. This will enable fast, accurate, and secure interpretation of next-generation sequencing to help as many people as possible get the right treatments.
