AWS for Industries
Part 2: Automated End-to-End Genomics Data Storage and Analysis using Amazon Omics
We would like to acknowledge the valuable contributions of David Obenshain, Gargi Singh Chhatwal, and Joshua Broyde, who are Solutions Architects at AWS.
With the rapid advancement and use of Genomic sequencing within research and clinical organizations, we need reliable, reproducible, scalable, and cost-efficient infrastructure to manage data and compute resources. Organizations want to spend less time on the undifferentiated heavy lifting of setting up and maintaining the infrastructure, and instead focus on their scientific business differentiators.
In part one of this blog post, we described the business challenges of storing and processing genomics and other biological data faced by healthcare and life sciences customers. We introduced Amazon Omics―a purpose-built service to specifically address these challenges. Amazon Omics will help bioinformaticians, researchers and scientists:
- Store raw and processed data
- Process their raw data through bioinformatics pipelines
- Query and analyze genomic variants to generate insights across cohorts of samples
In this blog post, we show how to use Amazon Omics with AWS Step Functions to automate the process of transforming raw sequence data to insights seamlessly. We describe an example reference architecture with sample code that can be used to automate the storage and analysis of incoming raw sequence data all the way to query-ready variants. Note this sample code is for demonstration purposes and not for production use.
Overview of Solution
This solution has been built with best-practices such as fine-grained permissions, Amazon Simple Storage Service (Amazon S3) lifecycle policies, and Infrastructure as Code using AWS CloudFormation. Users can customize these as needed.
The solution is designed to use serverless services and have minimal manual intervention, which reduces the operational burden for customers. We use AWS Step Functions and its native integration with AWS Lambda (Lambda) to automate the orchestration of tasks required to store, analyze and query genomics data with Amazon Omics.
We used an example workflow based on the GATK Best Practices from the Broad Institute to process raw genomic data to variants. We also used publicly available data required by this workflow from the GATK Resource Bundle.
The AWS CloudFormation template that is part of this solution, once deployed, creates the following AWS resources:
- S3 buckets with lifecycle rules for input raw sequence data and workflow output data, so that data is either transitioned to a lower tier or deleted after it is loaded into Amazon Omics Storage.
- AWS CodeBuild builds and deploys the necessary Lambda Functions and Docker Images (pushed to the account’s private Amazon Elastic Container Registry) required for automation.
- Amazon Omics resources necessary for data storage, secondary analysis workflow and variant data ingestion:
- Omics Reference Store and automated import of a reference genome.
- Omics Sequence Store.
- Omics Workflow and automated creation of an example Secondary Analysis workflow.
- Omics Variant Store (
omicsvariantstore) - Omics Annotation Store (
omicsannotationstore) with an automated import of ClinVar VCF.
- Step Functions workflows orchestrate data import into Omics Sequence and Variant Stores and an automated launch of the pre-created Omics Workflow.
- Lambda functions and Amazon S3 notifications trigger creation and launch of various resources.
 Figure 1: Reference architecture for an automated end-to-end solution to store and analyze Genomics data using Amazon Omics, AWS Step Functions and AWS Lambda
Figure 1: Reference architecture for an automated end-to-end solution to store and analyze Genomics data using Amazon Omics, AWS Step Functions and AWS Lambda
Once the resources are created, we are ready to process genomic input data in FASTQ format. Figure 1 shows the sequence of events as the raw data gets processed:
- Users upload raw sequence data in FASTQ format to the existing S3 bucket designated for inputs.
- The input S3 bucket has an Amazon S3 Event Notification that is configured to trigger a Lambda function. The Lambda function evaluates the file names and triggers the AWS Step Functions Workflow when a pair of FASTQs for the same sample name is detected.
- The AWS Step Functions Workflow includes steps that invoke AWS Lambda functions to:
- Import the FASTQs into an existing Omics Sequence Store.
- Start the pre-created Omics Workflow that uses GATK best practices Omics Workflow for Secondary Analysis.
- Once the workflow is complete, import workflow output BAM and VCF into the Omics Sequence and Variant Stores respectively.
- Tag input and output files in Amazon S3 so that they lifecycle into colder storage tiers or get deleted over time.
- The Variant store and Annotation store tables appear as resources in AWS Lake Formation.
Query Variant Data
Variant and Annotation data imported into Omics Variant Store and Annotation store are surfaced as shared tables using Omics’ integration with AWS Lake Formation (Lake Formation). This provides users with the ability to implement fine-grained access control. Using the sample code above, the omicsvariantstore and omicsannotationstore tables will appear in Lake Formation.
Lake Formation allows data lake Admins to grant AWS Identity and Access Management (IAM) users and roles access to desired databases and tables. Below we describe the steps that an Admin needs to make certain users have the right level of permissions to be able to query the data. Once the permissions are granted, users can access Variant and Annotation data using Amazon Athena (Athena).
Set up AWS Lake Formation permissions
The steps to grant the necessary permissions in Lake Formation to query the tables in Amazon Athena are as follows:
- Navigate to the AWS Lake Formation console and ensure that your IAM role is added as an Admin (see Figure 2).
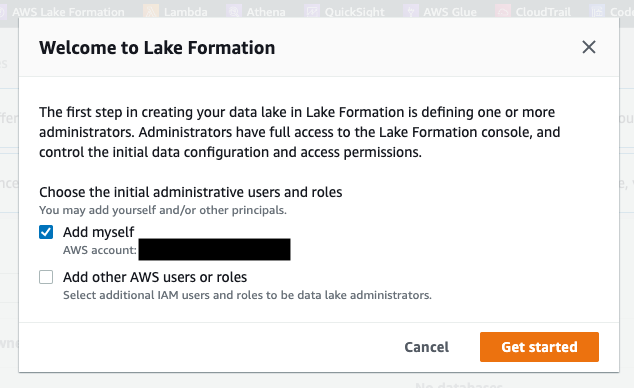 Figure 2: Add your IAM role as an administrator in AWS Lake Formation
Figure 2: Add your IAM role as an administrator in AWS Lake Formation
- On the “Create Database” screen click on “Database” on the left and create a database called
omicsdb. We will add the variant and annotation tables to this database (see Figure 3).
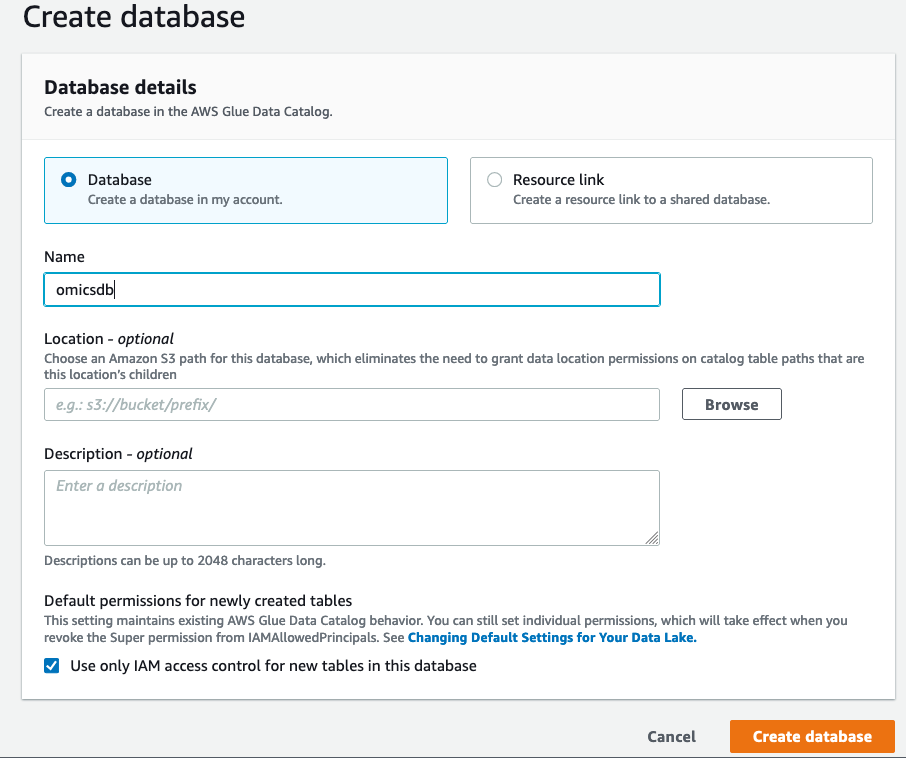 Figure 3: Create a database to add your variant and annotation tables
Figure 3: Create a database to add your variant and annotation tables
- Click on Tables on the left of the console. Select the
omicsvariantstoreand under “Actions”, select “Create Resource Link” (see Figure 4).
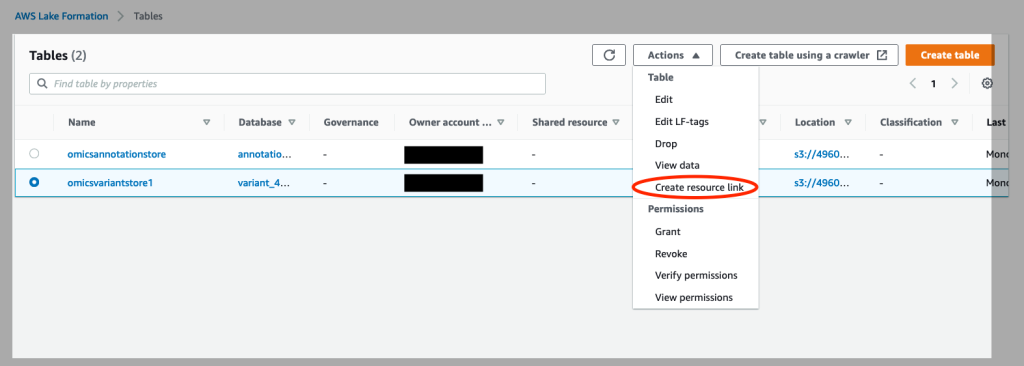 Figure 4: Create resource links for your Omics annotation store and Omics variant store in AWS Lake Formation
Figure 4: Create resource links for your Omics annotation store and Omics variant store in AWS Lake Formation
- On the “Create Resource Link” screen, give the resource link a name (for example,
omicsvariants) and choose the database you just created (omicsdb) in Figure 2.
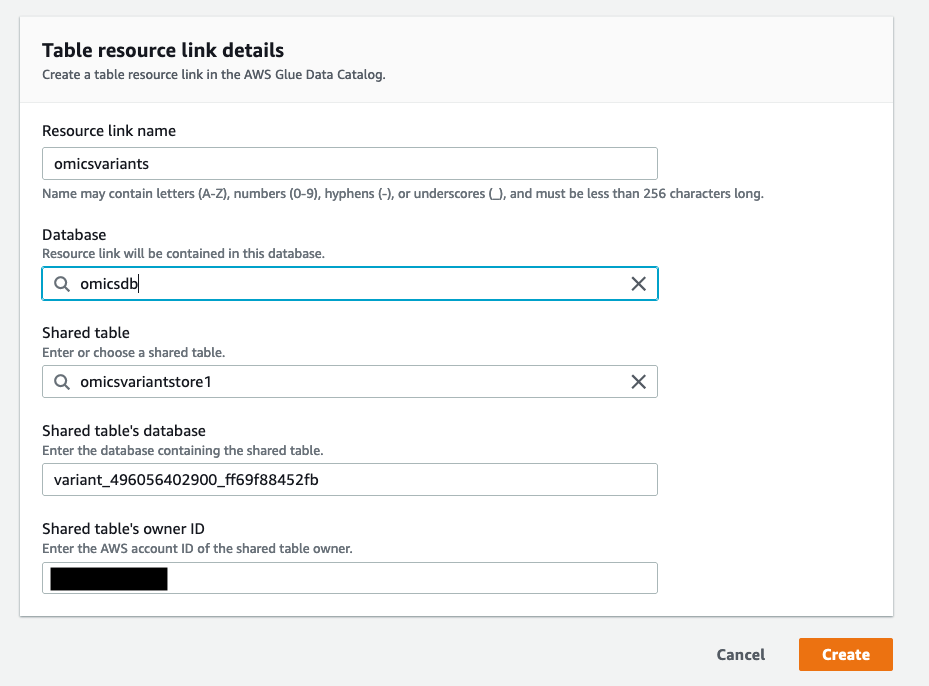 Figure 5: Create a resource link for the Omics Variant Store and add it to the newly created database
Figure 5: Create a resource link for the Omics Variant Store and add it to the newly created database omicsdb
- Repeat steps 3 and 4 for the creation of
omicsannotationstore.
Now that we have the variants and annotations available as a database in Amazon Athena, they can be queried through SQL queries in the Athena console. Note that the Athena workgroup for these queries needs to use Athena Version 3 query engine.
To query variants through an Amazon SageMaker (SageMaker) notebook, there is an additional step of granting permissions to the SageMaker notebook instance role to query the Variant and Annotation tables.
- Go to the AWS Lake Formation console and click on “Tables”.
- Choose the resource link for the variants we just in the AWS Lake Formation permissions.
- Under Actions, choose “Grant on Target”.
- On the “Grant Permissions screen”, choose the SageMaker execution rule that your notebook uses for the IAM principal.
- Under Table Permissions, choose “Select” and “Describe”.
- Leave all other selections in their default setting as shown in Figure 6.
- Repeat steps 1-6 as described here for Amazon SageMaker notebook instance roles for the
omicsannotationsresource link.
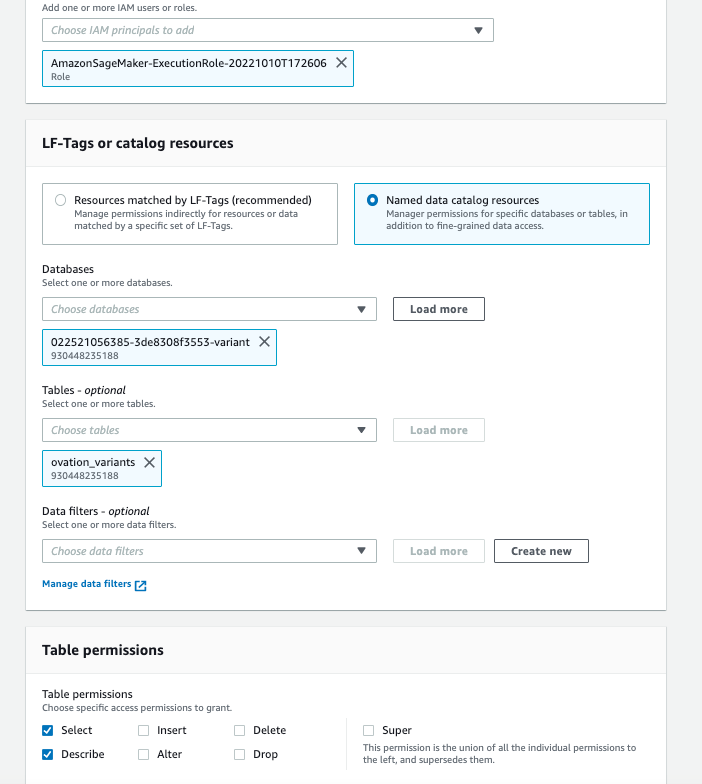 Figure 6: AWS Lake Formation permissions for the SageMaker Notebook execution role for the Omics variant and annotation resource links.
Figure 6: AWS Lake Formation permissions for the SageMaker Notebook execution role for the Omics variant and annotation resource links.
A Jupyter notebook with example queries is available here. This sample notebook uses the AWS SDK for Pandas to connect to Athena to make queries.
From variants to insights
Customers that leverage multiomic and multimodal data in their data lake can jointly query these datasets with the newly added genomic variants and derive insights. In this example, we used whole genome sequencing variants and patient demographic data from the example Ovation Dx non-alcoholic fatty liver disease (NAFLD) dataset that is available on the AWS Data Exchange. We used variant annotations from ClinVar to demonstrate how we can perform queries to gain insights from this cohort of 10 patients.
The 30X coverage whole genome sequencing data from the 10 patients is imported into the Omics Sequence store, and a WDL workflow based on GATK best practices is kicked off to process the raw data to variants. The generated variants are imported into the Variant store, the ClinVar VCF is imported into the Omics Annotation Store, both of which show up as tables in AWS Lake Formation.
The patient information is in the form of a CSV file that can be crawled by an AWS Glue Data Crawler and is made available as a table. Joining the patient information with variants and variant annotations, we can gain insights into the cohort of patients.
 Figure 7: An example of a visual representation of insights using genomics and clinical data
Figure 7: An example of a visual representation of insights using genomics and clinical data
For example, Figure 7 shows the genes that have the Pathogenic or Likely Pathogenic variants that cause the conditions by patient. It is interesting to note that in this cohort of patients, which have been diagnosed with NAFLD, all the patients have a variant in the APOA2 gene that increases their risk of Familial Hypercholesteremia. There is some evidence that there is a link between NAFLD and high cholesterol levels and obesity. Several of the patients also have variants that pre-dispose them to Type-1 Diabetes in this cohort.
This example shows how novel insights can be gained by combining genomic variants with clinical annotations and phenotypes, and how Amazon Omics streamlines this for genomic data.
Conclusion
In this blog post we demonstrated how to:
- Automate an end-to-end process of ingestion of raw sequence data into Omics Sequence Store.
- Analyze the data through a pre-defined workflow using Omics Workflows.
- Generate query-ready genomic variants using Omics Variant Store that can be used for downstream analysis with data from other modalities in a data lake.
In addition, we also show how genomic variant and annotation data stored in Amazon Omics can be jointly queried with clinical data in the user’s data lake using Amazon Athena and Amazon SageMaker.
With this architecture, customers can deploy a reproducible and automated end-to-end infrastructure that minimizes operational overhead and accelerates time to insights.
The sample code is available on GitHub. The README file in the GitHub repo has detailed instructions on how to set it up. Note this sample code is for demonstration purposes and not for production use.
You can check service availability in your region and pricing details at the Amazon Omics product page.
To know what AWS can do for you contact an AWS Representative.