AWS Partner Network (APN) Blog
Accelerating Genomics and High Performance Computing on AWS with Relevance Lab Research Gateway Solution
By Kiran Kumar Ballari, Principal Enterprise Technologist – AWS
By Robert M. Bata, Business Development Manager – AWS
By Puneet Chaddah, CTO; and Sundeep Mallya, Head of Research Gateway Product – Relevance Lab Inc.
 |
| Relevance Lab |
 |
Running genomics and high performance computing (HPC) workloads is complicated. It takes time to set up and get started. Instead of building their own solutions, researchers are always looking for simple and efficient ways to run their research and genomics workloads.
This means provisioning secure, performant, and scalable research environments with role-based access to workbench and domain-specific data, so they can focus on research rather than building and operating a solution on their own.
To address these customer asks, Relevance Lab developed Research Gateway—a solution that delivers secure and scalable research without customers having to do the heavy lifting. The solution enables customers to provision environments in minutes, allowing scientists to do just-in-time analysis with a selection of popular genomics analysis engines (for example, Illumina DRAGEN and NVIDIA Parabricks).
Pipeline processing is made easy with orchestrated workflow managers built on the Nextflow and Cromwell languages. Research Gateway also provides customers with a secure and resilient data ingestion framework and a complete view of costs, to help build budgets and plan for future workloads.
In this post, we provide an overview of the solution architecture and standard genomic research workflow. Focusing on a single use case, the post includes a walkthrough of how to access Research Gateway, provision products required for their genomics sequencing analysis, and run the analysis. We also demonstrate how to view the outputs and deep-dive on the cost components of this workload.
Relevance Lab is an Amazon Web Services (AWS) Partner and platform-led services company specializing in cloud, DevOps and automation, analytics, and digital transformation.
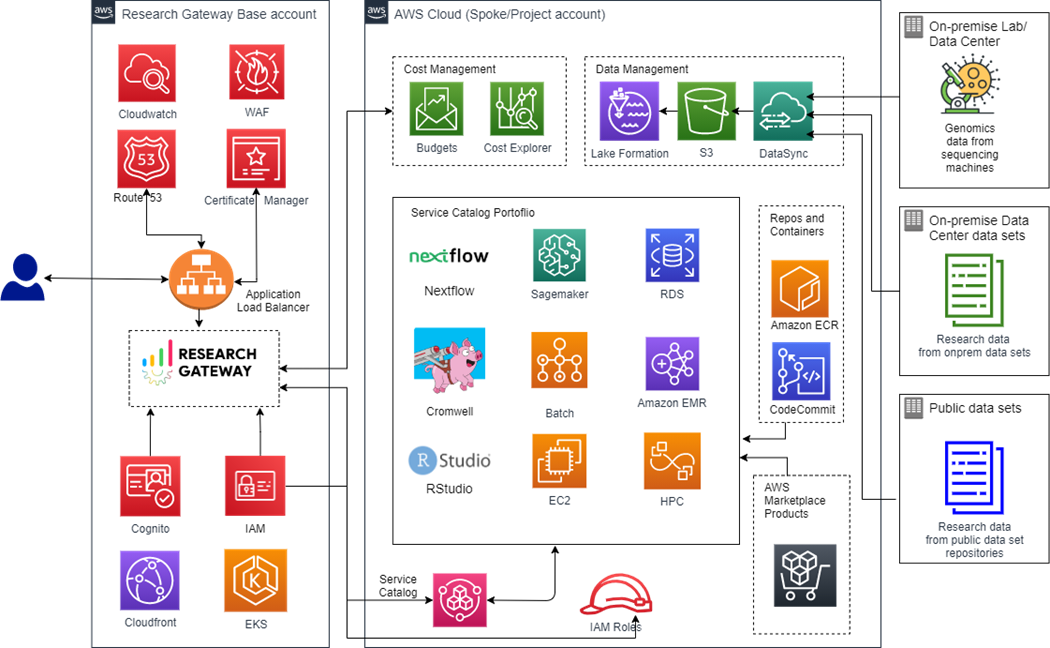
Research Gateway Architecture
Research Gateway architecture allows products to be provisioned in minutes in a simple interface that is easy to navigate and use by researchers. The Research Gateway instance is deployed using Amazon Elastic Kubernetes Service (Amazon EKS) and acts as the orchestration layer, providing capabilities to manage research projects.
Each project is managed in a separate AWS account to provide granular governance and control, based on the administrative unit.
Figure 1 – Research Gateway architecture.
The solution architecture uses core AWS services and open-source components to address common problems facing researchers, including:
- Access research environments in minutes, with user sign-up, authentication, and federation provided by Amazon Cognito and third-party identity providers. With a simple interface, administrators and lead researchers manage researcher permissions and accounts, as well as workspace permissions.
- Ease of access through self-service mechanism to deploy repeatable pre-built templates comprising infrastructure, application, and workflow components. A self-service catalog built on AWS Service Catalog is presented to end users to find and deploy approved IT templates and AWS Marketplace products.
- Secure access for users and administrators to access project resources using browser-based interfaces, secured by SSL. To make this process scalable and reliable, the solution uses AWS WAF, AWS Certificate Manager (ACM), and Application Load Balancer. Finally, Amazon CloudFront is used to improve performance by lowering the latency between the keyboard and the web server.
- Cost transparency and spend controls using AWS Budgets and AWS Cost Explorer to track projects, researchers, products, and pipelines. Consumption guard rails are implemented at the project level to flag any breach of project budgets and trigger pause or termination of the project.
- Common project storage, backed by Amazon Simple Storage Service (Amazon S3), for mounting genomics datasets for a project. This means customers can use public datasets hosted in AWS Open Data Sponsorship Program by default. Researchers can also bring on-premise large data sets on board using AWS DataSync or upload files using the drag and drop interface.
- Genomic workflow automation for secondary analysis using open source framework engines (Cromwell and Nextflow) and partner solutions (NVIDIA Parabricks and Illumina DRAGEN).
- Tertiary analysis with interpretation and deep learning using Genomics Data Lake integration, RStudio, and Amazon SageMaker.
- Reduction in time to research by selecting commonly-used nf-core and GATK pipelines that are pre-configured to execute on AWS Batch.
- Reuse of existing pipeline code by bringing their own code through AWS CodeCommit or containers from Amazon Elastic Container Registry (Amazon ECR).
- Environment monitoring, governance, and observability implemented through Amazon CloudWatch and AWS CloudTrail with the ability to integrate with customer enterprise systems.
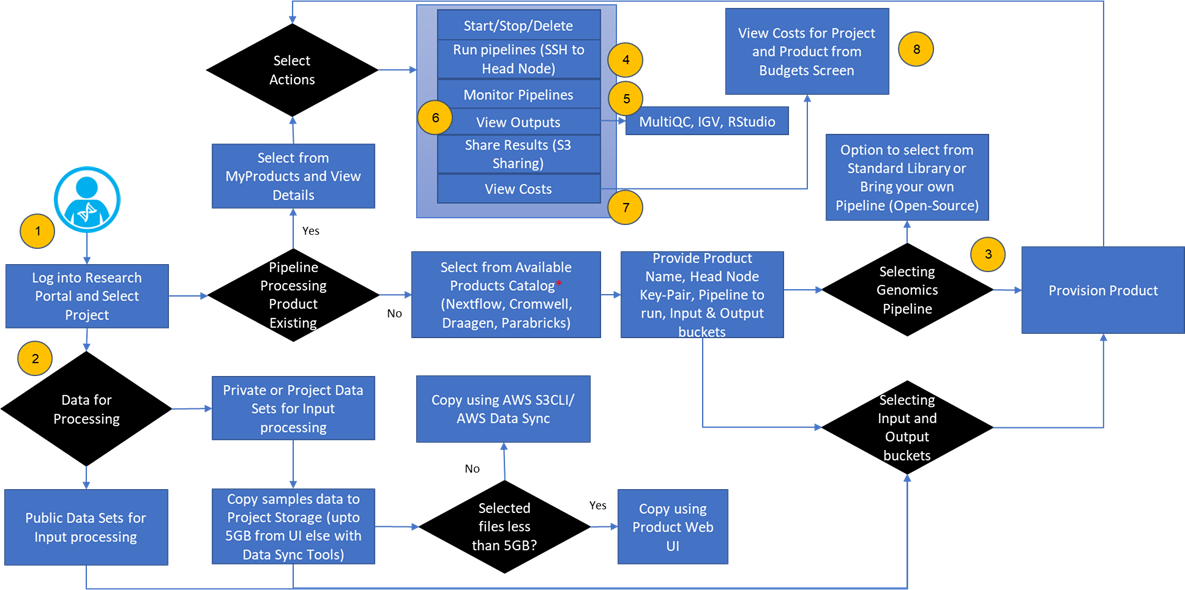
Genomic Sequencing Analysis Workflow
Genomics sequencing analysis using Research Gateway in the cloud enables researchers, scientists, developers, and analysts to efficiently run their experiments with no deep expertise in backend computing capabilities. The following walkthrough dives deep on the standard researcher workflow.
With Research Gateway, researchers can select reproducible workflows and standard public repository pipelines through the interface in users console and ingest raw data to input/output folders for sequencing analysis. Custom action triggers sequencing analysis steps, including quality checking, pre-processing, read alignment, variant calling, and annotation.
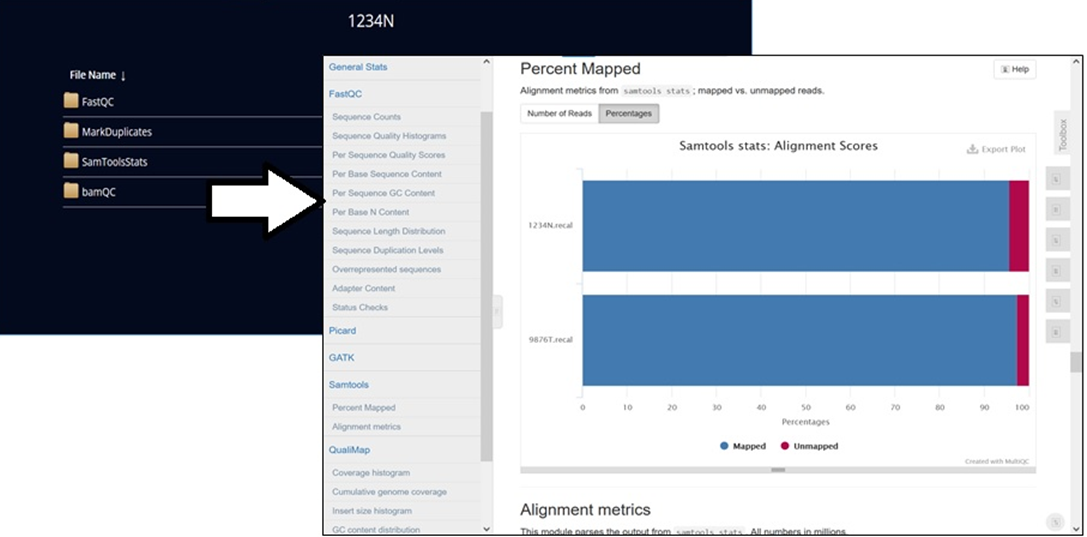
Researchers can visualize the processed output for the visual exploration and analysis of genomic data using interactive tools like Integrated Genomics Viewer (IGV), R-studio, and MultiQC.
Figure 2 – User workflow details.
While the solution allows any public pipeline built with Workflow Description Language (WDL), Common Workflow Language (CWL), and Nextflow specifications, this walkthrough uses Sarek as an example.
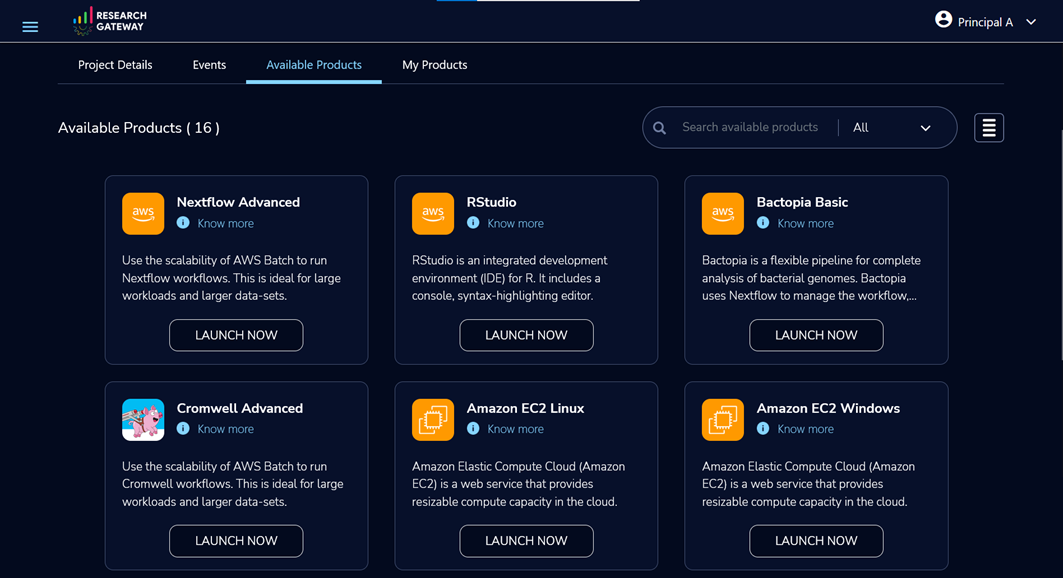
To get started, sign in to the Research Gateway portal with your login details. If you don’t have login details, select Sign up for new account. On the navigation page, choose Products to provision resources.
Figure 3 – Solution catalog of available products.
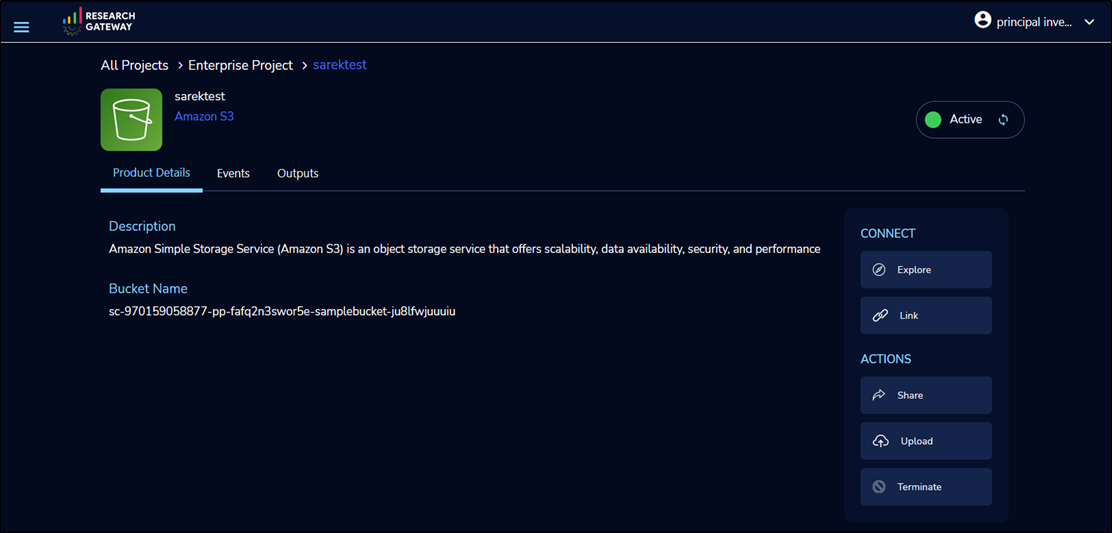
Next, Launch an Amazon S3 product to store and manage data.
To view the bucket contents, select Explore. To upload your input data to the bucket, select Add File/Add Folders.
In Product Details, copy the Bucket Name to the clipboard. You will need it later in the pipeline configuration.
Figure 4 – Amazon S3 bucket for storing data samples for genomic analysis.
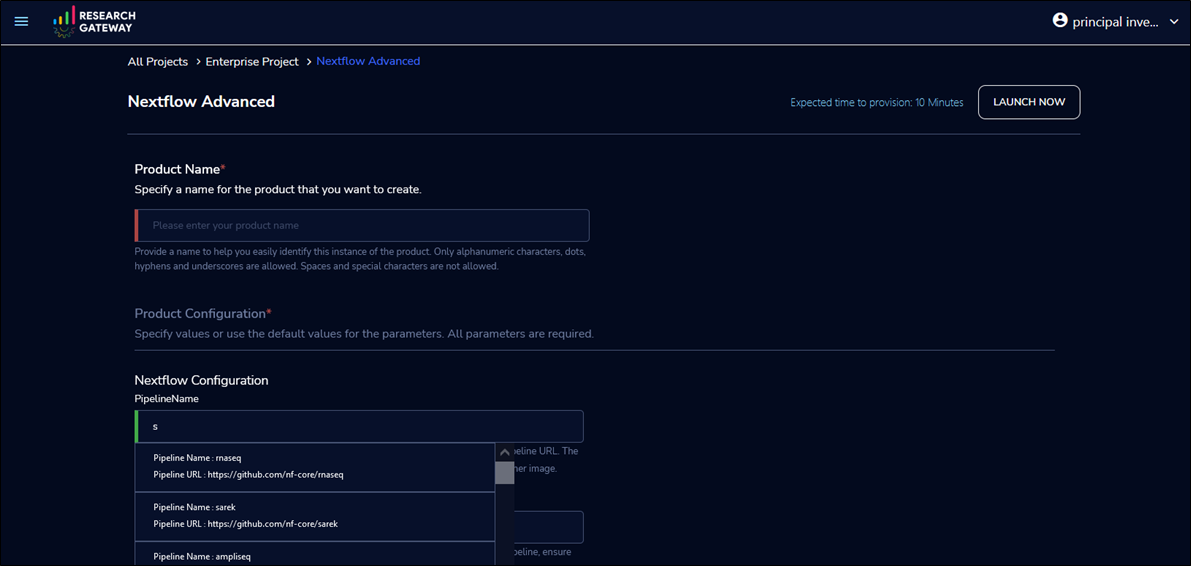
To execute the Sarek pipeline, launch a Nextflow Advanced product. Then select the nf-core/sarek pipeline in the PipelineName field by searching “Sarek”.
Pipeline configuration supports multiple instance types, Amazon S3 buckets, job submission queues, and job schedulers, like AWS Batch and Slurm Workload Manager through AWS ParallelCluster.
Figure 5 – Create a workspace for Nextflow choosing the Sarek pipeline.
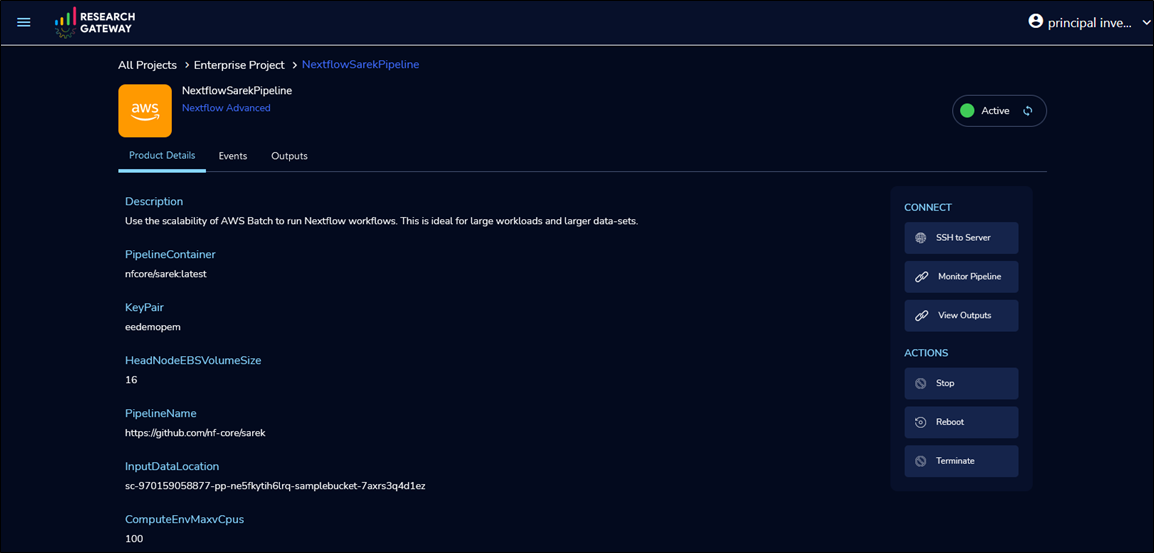
Once provisioning is complete, execute with default settings or select SSH to Server to connect to the head node and execute pipeline with custom configurations.
Figure 6 – Run the pipeline by securely connecting to the head node.
To monitor the job progress, select Monitor Pipeline. This launches the Nextflow Tower URL in a separate browser tab. Completed job stores MultiQC output in Amazon S3.
Figure 7 – Monitor the pipeline run using the integrated Nextflow Tower.
To view the output files, select View Outputs (as illustrated in Figure 6). Output generated automatically consists of variant call files and QC reports that are accessible to the user from the analysis environment. Customers can also download the files by clicking the links.
Figure 8 – View and download output files using the links.
The budgets and reporting dashboard provides the ability to configure and track cost transparency, alerts, and spend controls. These controls apply to projects, researchers, and products consumed in the Research Gateway.
Research Gateway provides a framework to integrate with customer charge back systems. Alternatively, customers can view and download cost reports. Personas selected by the customer—for example, administrator or principal researcher—have the ability to enforce pause and stop resources at project level if the consumed budget set exceeds threshold value.
Figure 9 – Monitor project, researcher, and resource level aggregated costs.
Summary
To help researchers provision secure, performant, and scalable research environments with role-based access to workbench and data, Relevance Lab developed the Research Gateway solution.
In this post, we walked through the steps a researcher would take to access Research Gateway, provision products required for genomics sequencing analysis, run the analysis, generate and view research output, and review the cost of the workload.
To get started with genomics research on AWS and to run your first genomic workload in less than 30 minutes, sign up for a new account.
To learn how Research Gateway can help customers accelerate research and how its functionality maps to customer use cases, contact the team at marketing@relevancelab.com.
Relevance Lab – AWS Partner Spotlight
Relevance Lab is an AWS Partner and platform-led services company specializing in cloud, DevOps and automation, analytics, and digital transformation.
Contact Relevance Lab | Partner Overview | AWS Marketplace
*Already worked with Relevance Lab? Rate the Partner
*To review an AWS Partner, you must be a customer that has worked with them directly on a project.