AWS for Industries
Deploy digital biomarkers at scale in clinical trials using serverless services on AWS
Background
Traditional assessments of disease progression and intervention efficacy used in clinical trials are often limited. These assessments normally consist of clinical outcome assessments (COAs) and patient reported outcomes (PROs). COAs are physician-derived, often requiring patients to travel to a clinical site. They are typically subjective, episodic in nature, and provide limited insight into the fluctuations of symptoms experienced outside the clinic [1]. PROs are multi-question surveys taken by patients, usually in their home environment. While PROs provide some insight into symptom severity from the patient’s perspective outside the clinic, they often suffer from recall bias, are impacted by mood or suggestion, and are qualitative in nature [2, 3]. Therefore, there is a need for more objective, continuous assessments of symptom severity outside the clinic to complement current assessment tools used today.
Wearable sensing technology has enabled many new approaches for objective, continuous assessments of patients during their daily lives. Over the past decade, researchers have leveraged wearable sensors to create digital biomarkers aimed at extracting clinically meaningful data about patients outside the clinic environment. Digital biomarkers are defined as objective, quantifiable physiological and behavioral data collected and measured using digital devices (e.g., smartphones and wearable devices). Recently, efforts have been made to use these digital biomarkers to measure disease progression and efficacy of interventions in clinical trials.
The need: Efficiently process incoming wearable sensor data from clinical sites around the world
The Digital Medicine and Translational Imaging (DMTI) group at Pfizer (part of Early Clinical Development within Worldwide Research, Development, and Medical) is developing and deploying digital biomarkers, derived from data captured using wearable devices in several large clinical trials. Participants in these trials wear devices containing accelerometers and near-body temperature sensors for a pre-determined monitoring period. After completing the monitoring period, participants return the devices to their respective clinical sites where the sensor data from each device is extracted and sent to Pfizer for processing.
The Pfizer DMTI team needed an efficient, scalable, and automated way to run their custom-built digital biomarkers (comprising of machine learning and heuristic, rule-based algorithms, all built in Python) on participant sensor data, and looked to AWS to build a solution. The following requirements had to be met when designing and building this solution:
- Scalability – The application must be scalable (Sample size ranging from dozens of participants (Phase 0/I) to thousands of participants (Phase III/IV)).
- Flexibility – The system should allow for ease of swapping in and out different digital biomarker solutions based on the respective use case of interest.
- Reproducibility – The digital biomarker solutions need to be immutable as they are run within the application to ensure consistent results.
- Quality/Security – The system must adhere to key regulatory standards and research practices pertaining to data privacy and security. This includes, as applicable, GxP and FDA Title 21 CFR Part 11 compliance, among others.
Designing the infrastructure
Pfizer DMTI Data Scientists Yiorgos and Nikhil collaborated with AWS Solutions Architect Michael to take a simplistic approach when designing the infrastructure to process sensor data as described above. The core concepts they wanted to include were:
- Infrastructure-as-code – to allow for easy management of infrastructure stack as well as ability to easily take down and re-build stack from code
- Serverless – to reduce burden of server management and allow for faster experimentation during the development phase
- Data storage – to house incoming raw sensor data and intermediate data outputs during pipeline processing
- Event-based – to enable automated start of the pipeline when data is present
- Compute engine – to run Python-based digital biomarker solutions on raw sensor data
- Algorithm storage – to house specific versions of digital biomarker solutions
- Monitoring and observability – to appropriately alert team about run-time issues that may occur within the pipeline
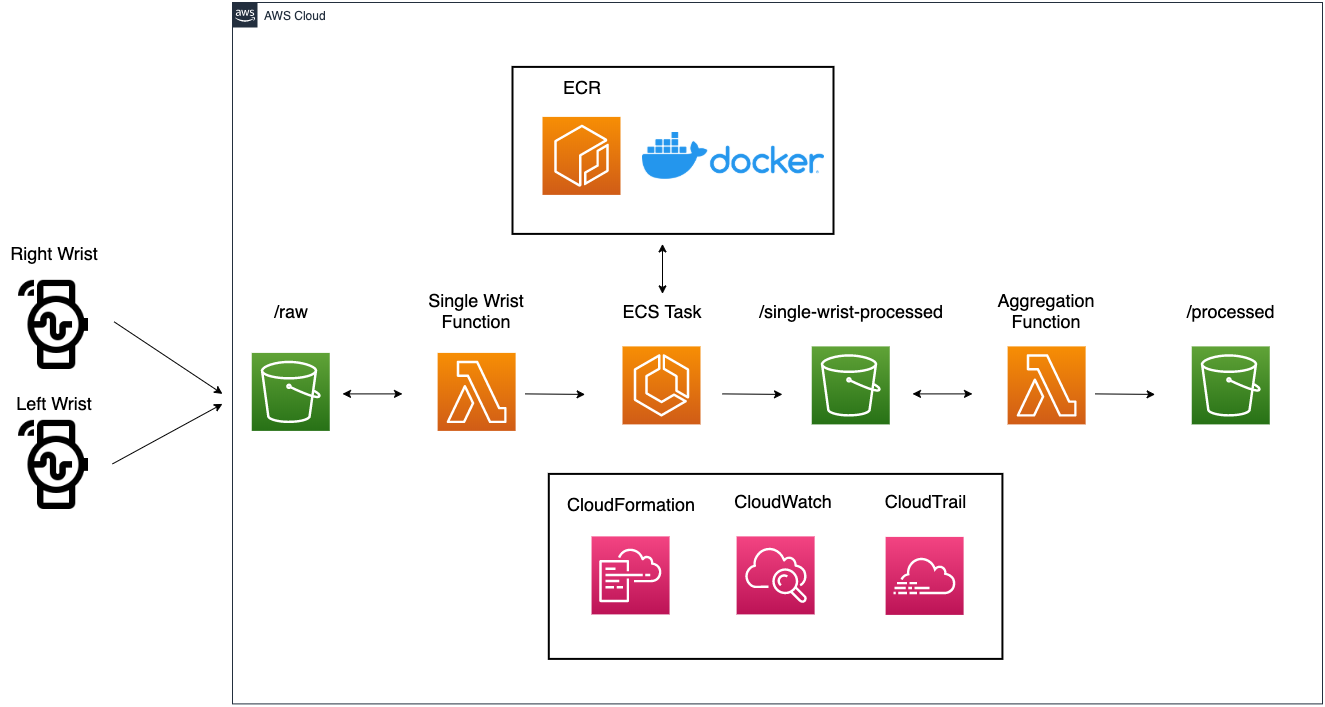
The solution architecture
Before delving deeper, let’s walk through each step of this pipeline:
- Incoming sensor data (.CSV files) from wearable devices worn on both wrists are saved in an Amazon S3 bucket
- An AWS Lambda function is subscribed to a S3 PUT event. The single wrist Lambda function then requests an Amazon Elastic Container Service (ECS) task with a pointer to the newly uploaded data file.
- Digital biomarker solutions are stored as Docker images housed in Amazon Elastic Container Registry (ECR). Once ECS receives RunTask from the single wrist Lambda function, it pulls the Docker image of interest from Amazon ECR, spins up a container with the digital biomarker solution installed, and runs the digital biomarker solution on the respective data file.
- After the run is complete, intermediate outputs (results from the respective wrist data file) are saved back into S3 as CSV files under /single-wrist-processed prefix.
- The aggregation function listens for intermediate outputs saved from both wrists (parsing file names saved in the S3 bucket; left and right wrist as well as random participant identifier are denoted in filename). Once both outputs are saved in S3, the Lambda function downloads both files (outputs from right wrist and left wrist), and aggregates into a final set of outputs for the given participant.
- These final outputs are saved back into the S3 bucket, under /processed prefix.
Building the stack with AWS CloudFormation
They leveraged AWS CloudFormation to provision and configure all resources for this architecture. This provides the ability to easily add and remove services while going through the development phase, as well as version controlling the infrastructure-as-code in Pfizer’s local GitHub. Adding CloudFormation templates to version control was an integral part of the change management solution deployment and audit documentation.
AWS Command Line Interface (CLI) was used to upload new versions of the CloudFormation template as they iterated. (For instructions, see Installing AWS CLI and Configuring the AWS CLI). Using CLI and bash scripting removes manual steps, which reduces the time to deploy and potential for human error. An example of the commands used to upload the template using the AWS CLI can be seen in the bash script below:
Building the Docker image
By using Docker, the Pfizer team can treat each digital biomarker solution as a module that can be easily swapped in and out based on the use case. With ECS and Docker, they can generalize the pipeline and swap the digital biomarkers as needed, allowing them to scale the pattern beyond this use case. The digital biomarker solution code and all of its dependencies are installed in the Docker file. The Docker file pulls directly from private GitHub repositories with pointers to tagged releases, ensuring proper versioning when deploying. (This requires having GitHub SSH setup; see Setting up GitHub SSH). This secure integration with GitHub protects any proprietary code or data while still allowing the automated retrieval and installation of all necessary software by the pipeline. See the example Docker file:
Then, the compiled Docker image is uploaded to AWS ECR with the AWS CLI and bash:
Leveraging AWS Lambda and Amazon ECS for event-driven processing
As mentioned in steps two and three of the infrastructure architecture, AWS Lambda was used to initiate the pipeline by listening for upload events and routing the associated file information to Amazon ECS via a handler function. This event-driven approach allowed for full automation of the pipeline and any data upload or transfer cadence desired, and played a decisive role in facilitating parallel processing.
Monitoring pipeline runs with Amazon CloudWatch
A standardized logging structure was created so that dashboards could be built in Amazon CloudWatch, which parse the output log files for issues or errors during each pipeline run. The logging format comprised of a series of tags ([ERROR], [PIPELINE_ISSUE], [ALGORITHM_ISSUE], etc.). These tags were then parsed and displayed along with other relevant metrics (Lambda invocations, number of files in bucket, etc.) on a custom-built CloudWatch dashboard to allow for monitoring of pipeline runs. An example of the dashboard is shown in the following image:
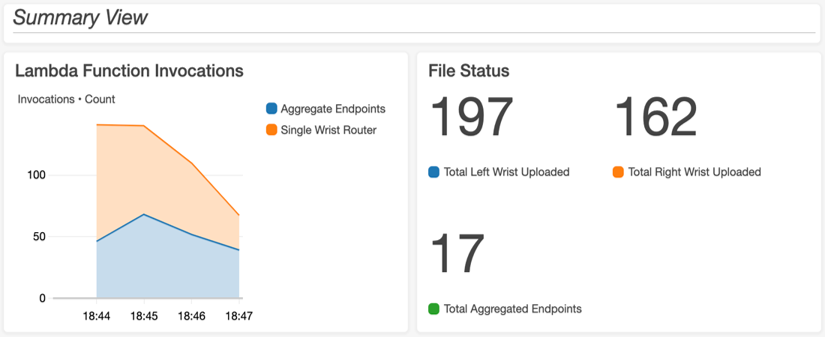

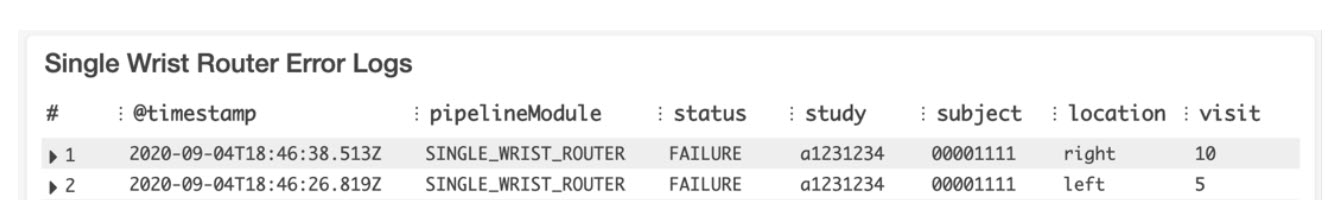
Each log tile uses CloudWatch Logs Insights with a custom query to parse all the log files in the log stream for the tags of interest. The Single Wrist Router Error Logs were generated by the following example query:
Load testing
While developing the pipeline architecture, small test files were used for quick feedback on how the build was progressing. However, the final pipeline is meant to handle hundreds or even thousands of files coming in from sites all over the world. To pressure test the system and ensure that the logging structure and monitoring schema could handle real-world scenarios, load testing was performed in various ways. The first test uploaded 250 pairs of left/right sample data to ensure that a large data dump would be handled correctly. Data should be recognized and distributed to individual compute instances for each file in parallel, intermediate data should trigger the appropriate aggregation step for the left and right wrist of each pair of input files regardless of upload order, and the relevant run metrics and logs should appear and effectively communicate run status in the dashboard. Further testing was performed again with hundreds of files with real data available from their research studies. This latter set of tests were used to determine how gracefully the pipeline could handle real world data and any resulting processing failures, such as missing data, data from devices that were not worn, and mislabeled data.
Key learnings
Using an iterative development strategy, the Pfizer DMTI team and AWS broke down each feature development process into small work packages with small sample datasets to test each new feature, with the goal of having a working solution after each iteration. This let them quickly isolate and fix errors, progressing forward in a methodical but fast manner.
Using Docker along with AWS ECS/ECR helped achieve target goals of flexibility and reproducibility. Having each digital biomarker solution wrapped in a Docker image with standardized input and output designs let the Pfizer DMTI team easily swap in and out different digital biomarker solutions. This approach also met reproducibility requirements: digital biomarker solutions were completely locked down, ensuring that the digital biomarker solution was run in the same manner every time the application ran.
With the use of event-based architecture powered by Lambda and ECS, results could be automatically processed. This lets the Pfizer DMTI team scale from small-scale studies of hundreds of participants to large-scale studies of thousands of participants without changing the architecture. Additionally, there would not be idle resources in between studies.
Lastly, as the goal was to process real clinical trial data, the team needed to meet various security, quality, and compliance standards. By following AWS best practices and the AWS Shared Security Model, it was easy to implement least privilege (users only access resources necessary for users’ purpose) within the application and meet security goals. Partnering with Pfizer’s Cloud Platform Team, who qualified AWS services for GxP compliance, ensured a path to validate the architecture for GxP validation. CloudWatch and CloudTrail were instrumental in capturing all events happening in the pipeline, satisfying key audit requirements around user roles and error monitoring.
Conclusions & next steps
With cloud computing on AWS, the Pfizer team was able to run digital biomarker solutions at scale across clinical trials with ease. Here are the three key benefits they found when using AWS cloud computing for this task:
- High quality – By using services like Docker with Amazon ECS and Amazon ECR, they were able to maintain reproducibility, which is essential in regulated environments. Also, they were able to meet key regulatory standards and research best practices with this system.
- Time saving – Using AWS robust parallel processing capabilities enables significantly faster processing times. For example, they processed ~36,000 hours of data in about 20 minutes, which is significantly faster than the seven to 12-day run-time estimated for on-premises processing.
- Cost saving – AWS has pay-as-you-go pricing models, so processing tasks cost far less than managing internal servers and personnel to handle the processing loads.
By using AWS services, the Pfizer team could make updates quickly after the initial deployment. Next steps include expanding the number of digital biomarker solutions that are supported today and integrating services like AWS Step Functions so that they can further simplify the architecture.
AWS welcomes your feedback. Feel free to leave questions or comments in the comment section on this post.
—
References
- Dorsey, E. R., Venuto, C., Venkataraman, V., Harris, D. A. & Kieburtz, K. Novel methods and technologies for 21st-century clinical trials. JAMA Neurol. 72, 582 (2015).
- Papapetropoulos, S. S. Patient Diaries As a Clinical Endpoint in Parkinson’s Disease Clinical Trials. CNS Neurosci. Ther. 18, 380–387 (2012).
- Murray, C. & Rees, J. Are Subjective Accounts of Itch to be Relied on? The Lack of Relation between Visual Analogue Itch Scores and Actigraphic Measures of Scratch. Acta Derm. Venereol. 91, 18–23 (2011).
—
Guest authored by

Nikhil Mahadevan is a Manager of Digital Health Data Science in the Digital Medicine & Translational Imaging group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. He is responsible for leading design, development, and deployment of digital biomarkers used as new measures of disease progression in Pfizer’s clinical trials. Nikhil holds a Bachelor of Science degree in Biomedical Engineering from Boston University.

Yiorgos Christakis is a Digital Health Data Scientist in the Digital Medicine & Translational Imaging group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. His primary focus is the design, development, and deployment of digital biomarkers for monitoring disease progression in various populations for Pfizer’s clinical trials. Yiorgos holds a Bachelor of Science degree in Biomedical Engineering from Boston University and a Master of Science degree in Computer Science from Georgia Institute of Technology.