AWS Machine Learning Blog
Map clinical notes to the OMOP Common Data Model and healthcare ontologies using Amazon Comprehend Medical
Being able to describe the health of patients with observational data is an important aspect of our modern healthcare system. The amount of quantifiable personal health information is vast and constantly growing as new healthcare methods, metrics, and devices are introduced. All of this data allows clinicians and researchers to understand how the health of a patient changes over time and identify opportunities for precision treatment. The aggregate of this data informs epidemiologists about the health of populations and enables the identification of cause and effect patterns.
Unstructured text, commonly in the form of clinical notes, is a rich source of patient observational health data. Often, there is important information written by clinicians into notes that isn’t encoded into a patient’s structured health record. Clinical notes can also be used to help validate the quality of structured health record data. Historically, the challenge with clinical notes has been that they require manual review to extract the contained medical insights, which is time consuming and expensive.
Amazon Comprehend Medical is a natural language processing (NLP) service that uses machine learning to extract insights like medical condition, medication, dosage, and strength quickly and accurately. Customers can use Amazon Comprehend Medical in a pay-as-you-go model and immediately begin extracting insights from medical text without having to develop or train a complex machine learning model themselves.
The Observational Medical Outcomes Partnership (OMOP) Common Data Model, maintained by the Observational Health Data Science and Informatics (OHDSI) community, is an industry standard, open source data model used for health data. OMOP uses standardized medical ontologies, or “vocabularies,” like SNOMED, to store observational health data. From the OHDSI website:
“The OMOP Common Data Model allows for the systematic analysis of disparate observational databases. The concept behind this approach is to transform data contained within those databases into a common format (data model) as well as a common representation (terminologies, vocabularies, coding schemes), and then perform systematic analyses using a library of standard analytic routines that have been written based on the common format.”
A feature of OMOP is the ability to store clinical notes captured from disparate health data sources. In the data model, these notes are linked to an individual patient and a visit, giving context for the note. OMOP also has the ability to store insights inferred from notes by a natural language processing (NLP) engine. In this blog post we’ll explore how you can use Amazon Comprehend Medical to read notes from OMOP, extract medical insights, and write them back into OMOP using SNOMED ontological codes to enhance patient and population observational health data.
OMOP note processing architecture
This example works with clinical notes in the larger context of a full OHDSI architecture. You can learn more about our automated architecture for OHDSI on AWS by visiting this GitHub repository. The code given here specifically reads notes from and writes insights to the OMOP common data model. However, the same general process can be used with other structured data models for observational data that support clinical notes.
A primary feature of OMOP is the ability to consolidate data from many sources, like Electronic Health Record (EHR) systems and administrative claims data, from many geographic regions, into a common data model. In OMOP, the analysis of observational data from disparate sources is enabled by mapping dozens of source vocabularies (sometimes called dictionaries or ontologies) like SNOMED, RxNorm, ICD10, Read, LOINC, and others to an OMOP standard vocabulary. This enables OMOP to act as a kind of Rosetta stone to allow the interpretation of observational data between different sources and from different global regions.
One important aspect of the approach demonstrated here is mapping the entities detected by Amazon Comprehend Medical to the SNOMED ontology. Because observations must be expressed using standard codes in the OMOP data model, we must first map the insights detected by Amazon Comprehend Medical to a standard ontology before we can write them to OMOP. This is accomplished by sending entity text to the SNOMED CT Browser service to receive a SNOMED code for each entity. The following diagram shows the architecture for this process.
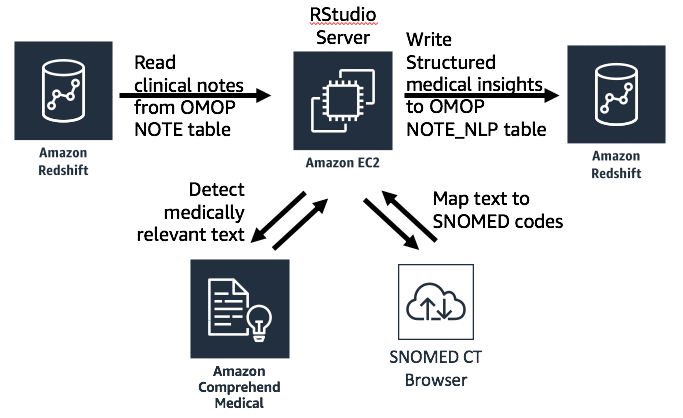
Using R code for notes processing
You can download the R Code demonstrating this process from GitHub.
This R code is demonstrated inside of the OHDSI on AWS architecture running on an Amazon EC2 instance running RStudio Server Open Source Edition instance. RStudio is a web-based, integrated development environment (IDE) for working with R. It can be licensed either commercially or under AGPLv3.
The code provided here can also be used from an environment outside of AWS to call Amazon Comprehend Medical. If you want to use it outside of AWS, you’ll need to configure a credentials file that allows this code to call the Amazon Comprehend Medical service. You can find instructions for doing that here.
At the very beginning of the notebook, you’ll see the following statements that define your OMOP CDM connection details and schema name. If you are using the OHDSI on AWS architecture, these are provided for you in a file called connectionDetails.R in your home directory. If outside of AWS, populate these lines with the details specific to your architecture.
Amazon Comprehend Medical returns a confidence score for every entity, trait, and attribute that it detects. Next in the code, you’ll see a variable that defines a minimum confidence score that detections must meet in order to be written to the NOTE_NLP table. This minimum confidence score can be altered as appropriate for your use case.
The actual calls to Amazon Comprehend Medical are implemented using the AWS SDK for Python (boto3). An R library called “reticulate” is used to execute this Python code from within R, pass in the note text, and receive back the entity data detected by Amazon Comprehend Medical.
Every note found in the OMOP NOTE table is read using a SQL query and processed. For each entity detected in each note by Amazon Comprehend Medical, an appropriate SNOMED code is determined by calling the SNOMED CT Browser.
As each note is processed you’ll see output to the R console that shows each record written to the NOTE_NLP table.
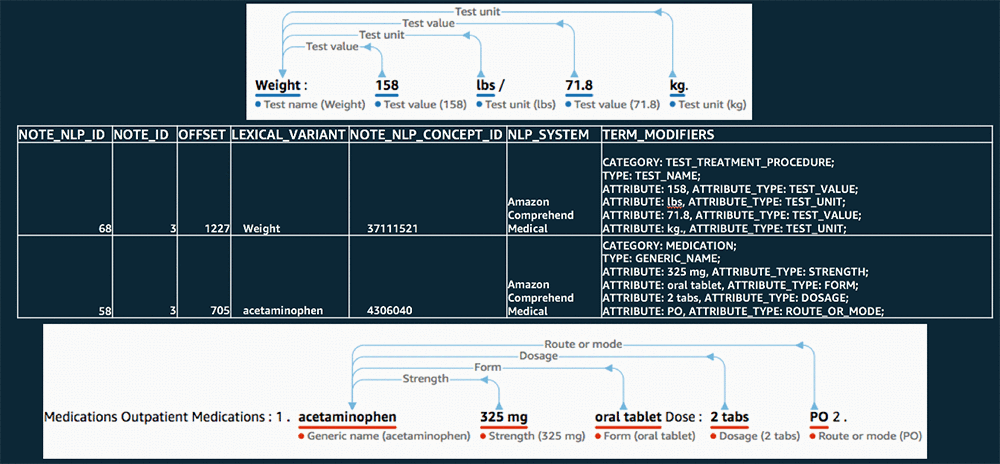
After the processing is complete, you’ll have a NOTE_NLP table filled with records that represent medical insights extracted from the notes in your NOTE table. Each of these records has an OMOP Standard Concept ID (the NOTE_NLP_CONCEPT_ID field), mapped from a SNOMED code, that represents the primary entity detected. The TERM_MODIFIERS field in the NOTE_NLP table is used to capture the category, type, and any attribute or trait data that Amazon Comprehend Medical detected about each entity.
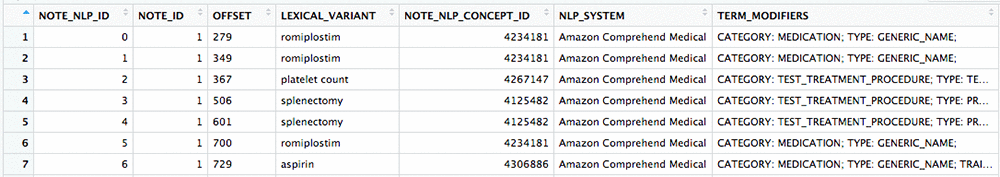
The following image shows a comparison of some original portions of note text with entities detected by Amazon Comprehend Medical and the corresponding NOTE_NLP record written to OMOP.
Conclusion
This example shows how Amazon Comprehend Medical makes natural language processing for medical text easily accessible. The insights detected by Amazon Comprehend Medical enable data scientists, epidemiologists, and medical researchers to extend patient and population structured observational health records with information that is locked away in unstructured notes. This additional data provides important insight for precision medicine, longitudinal studies, clinical trial candidate assessment, and population health studies. The code given here can help you get started processing notes into the OMOP Common Data Model today, or it can be used as a pattern to process clinical notes into any observational health data model.
About the Author
 James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.
James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.