AWS Machine Learning Blog
Using DeepChem with Amazon SageMaker for virtual screening
Virtual screening is a computational methodology used in drug or materials discovery by searching a vast amount of molecules libraries to identify the structures that are most likely to show the target characteristics.
It is becoming a ground-breaking tool for molecular discovery due to the exponential growth of available computer time and constant improvement of simulation.
Deep learning technologies are widely used in this computational virtual compound screening, and such technologies have evolved tremendously. DeepChem is one of the most popular open-source tools that democratizes the use of deep learning in drug discovery, materials science, quantum chemistry, and biology. For more information, see Democratizing Deep-Learning for Drug Discovery, Quantum Chemistry, Materials Science and Biology on GitHub.
This post describes how to use DeepChem with Amazon SageMaker. Amazon SageMaker is a fully managed service that enables you to quickly and easily build, train, and deploy machine learning (ML) models. ML often feels harder than it should because the process to build, train, and deploy models into production is complicated and slow. Amazon SageMaker removes that complexity.
Installing DeepChem with Amazon SageMaker
To install DeepChem, set up an AWS account and create your first Amazon SageMaker notebook instance. Complete the following steps:
- Create an AWS account if you do not already have one.
When you sign up for AWS, your AWS account is automatically signed up for all AWS services, including Amazon SageMaker. You are charged only for the services that you use. If you are a first-time user of Amazon SageMaker, see How Amazon SageMaker Works.
- On the Amazon SageMaker console, choose Notebook instances.
- Choose Create notebook instance.
- Under Notebook instance settings, for Notebook instance name, enter a name for your instance. This post uses the name
deepchem. - For Notebook instance type, enter your preferred type. This post uses
mL.t3.xlarge. - For Elastic Inference, choose None.
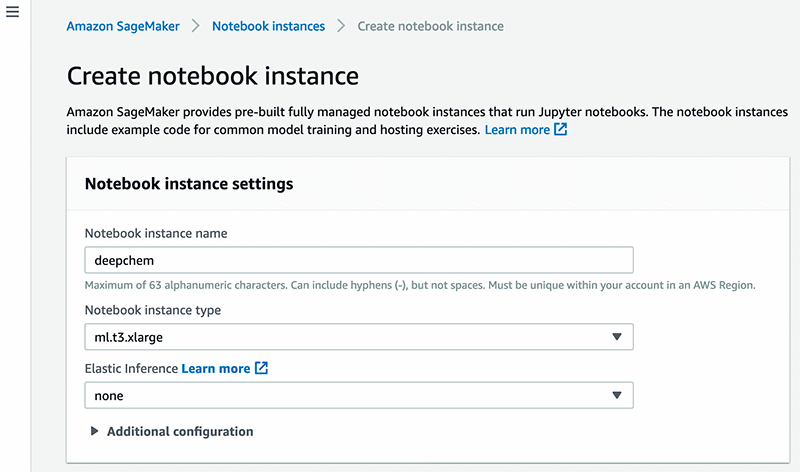
For more information, see Step 2: Create an Amazon SageMaker Notebook Instance.
When you finish the preceding steps to create a notebook instance, Amazon SageMaker launches an ML compute instance. The notebook instance has a preconfigured Jupyter notebook server and a set of Anaconda libraries.
- When the notebook instance status is
InService, choose Open Jupyter.
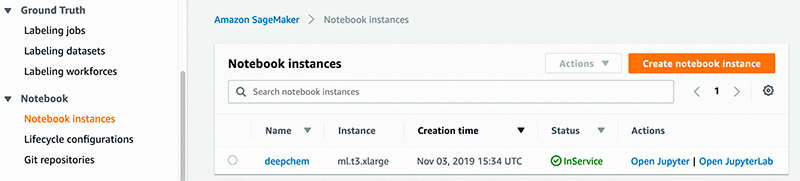
The Jupyter notebook server page appears.
- From the New drop-down menu, choose Terminal.

- Update Conda with the following code in your terminal:
You are now ready to install DeepChem.
- Run the following codes in your terminal:
- Create and check your DeepChem Notebook kernel with the following code:
- Close the terminal and Jupyter home window.
It can take up to a few minutes for Jupyter to recreate its drop-down menu.
Testing DeepChem installation
To test your DeepChem installation, complete the following steps:
- On the Amazon SageMaker console, choose Open Jupyter.
- On the Jupyter dashboard, from the New drop-down menu, choose
conda_deepchem.
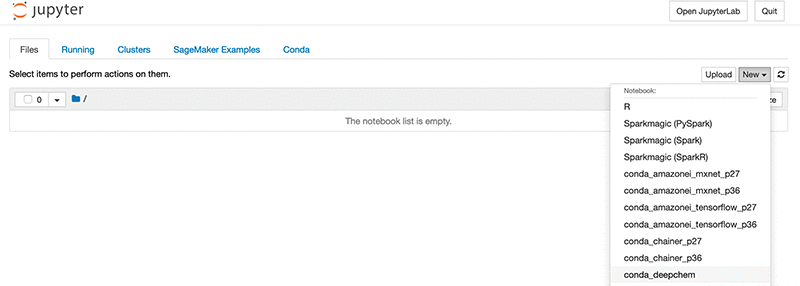
To check that DeepChem installed correctly, enter the following code into the notebook cell:
Running DeepChem
Now you can use DeepChem with the Jupyter Notebook App on an Amazon SageMaker notebook instance. The following example Python code generates uncertainty estimates. For more information, see Uncertainty in Deep Learning. You can load a dataset, create a model, train it on the training set, and predict the output on the test set. See the following code:
You see the following output and can check the prediction results in y_pred and y_std:
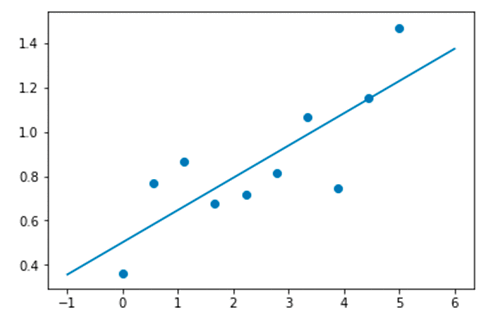
In this second example, the following code shows the best fit linear regression to a set of 10 data points:
You can see the following graph as the result.
Conclusion
This post demonstrated how to install DeepChem to Amazon SageMaker and verify it by running tests provided on the DeepChem Github repository. You can run your own codes or experiment with further DeepChem tutorials. For more information, see Tutorials on the DeepChem Github repository. The tutorials show off various aspects or capabilities of DeepChem. They can be run interactively in Jupyter (IPython) notebook. You can also download the notebook files and open them in the Amazon Sagemaker.
If you have any questions, please leave them in the comments.
About the Author
 Seongik Hong is a cloud infrastructure architect on the shared delivery teams. In his role, he leverages his experience to help people bring their ideas to life utilizing services provided by AWS. In his spare time, he enjoys experimenting how cloud computing can help cheminformatics.
Seongik Hong is a cloud infrastructure architect on the shared delivery teams. In his role, he leverages his experience to help people bring their ideas to life utilizing services provided by AWS. In his spare time, he enjoys experimenting how cloud computing can help cheminformatics.