AWS Big Data Blog
Perform biomedical informatics without a database using MIMIC-III data and Amazon Athena
Biomedical researchers require access to accurate, detailed data. The MIT MIMIC-III dataset is a popular resource. Using Amazon Athena, you can execute standard SQL queries against MIMIC-III without first loading the data into a database. Your analyses always reference the most recent version of the MIMIC-III dataset.
This post describes how to make the MIMIC-III dataset available in Athena and provide automated access to an analysis environment for MIMIC-III on AWS. We also compare a MIMIC-III reference bioinformatics study using a traditional database to that same study using Athena.
Overview
A dataset capturing a variety of measures longitudinally over time, across many patients, can drive analytics and machine learning toward research discovery and improved clinical decision-making. These features describe the MIT Laboratory of Computational Physiology (LCP) MIMIC-III dataset. In the words of LCP researchers:
“MIMIC-III is a large, publicly available database comprising de-identified health-related data associated with approximately 60K admissions of patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. …MIMIC supports a diverse range of analytic studies spanning epidemiology, clinical decision-rule improvement, and electronic tool development. It is notable for three factors: it is publicly and freely available, it encompasses a diverse and large population of ICU patients, and it contains high temporal resolution data including lab results, electronic documentation, and bedside monitor trends and waveforms.”
Recently, the MIT Laboratory of Computational Physiology (LCP) started hosting the MIMIC-III dataset on the AWS cloud through the AWS Public Dataset program. You can now use the MIMIC-III dataset via S3 without having to download, copy, or pay to store it. Instead, you can analyze the MIMIC-III dataset in the AWS Cloud using AWS services like Amazon EC2, Athena, AWS Lambda, or Amazon EMR. AWS Cloud availability enables quicker and cheaper research into the dataset.
Services like Athena also offer you new analytical approaches to the MIMIC-III dataset. Using Athena, you can execute standard SQL queries against MIMIC-III without first loading the data into a database. Because you can reference the MIMIC-III dataset hosted by MIT LCP in Amazon S3, your analyses always reference the most recent version of the MIMIC-III dataset. Live hosting reduces upfront time and effort, eliminates data synchronization issues, improves data analysis, and reduces overall study costs.
Transforming MIMIC-III data
Historically, the MIMIC team distributed the MIMIC-III dataset in compressed (gzipped) CSV format. The choice of CSV format reflects the loading of MIMIC-III data into a traditional relational database for analysis.
In contrast, the MIMIC team provides the MIMIC-III dataset on Amazon S3 in the following formats:
- The traditional CSV format
- An Apache Parquet format optimized for modern data processing technologies such as Athena
Apache Parquet stores data by column, so queries that fetch specific columns can run without reading the whole table. This optimization helps improve the performance and lower the cost of many query types common to biomedical informatics.
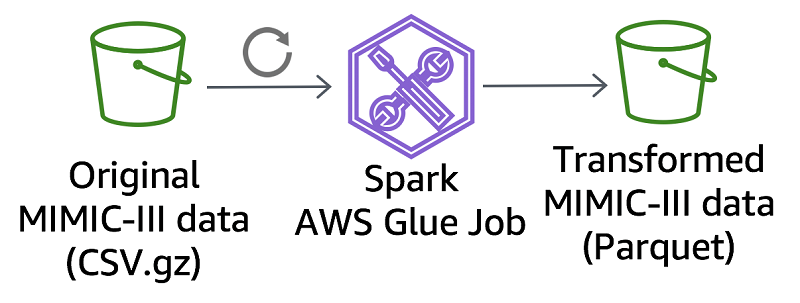
To convert the original MIMIC-III CSV dataset to Apache Parquet, we created a data transformation job using AWS Glue. The MIMIC team schedules the AWS Glue job to run as needed, updating the Parquet files in Amazon S3 with any changes to the CSV dataset. These scheduled updates keep the Parquet files current without interfering with the MIMIC team’s CSV dataset creation procedures. Find the code for this AWS Glue job in the mimic-code GitHub repo. Use the same code as a basis to develop other CSV-to-Parquet conversions.
The code that converts the MIMIC-III NOTEEVENTS table offers a valuable resource. This table stores long medical notes made up of multiple lines, with commas, double-quotes, and other characters that can be challenging to transform. For example, you can use the following Spark read statement to interpret that CSV file:
Executing the indwelling arterial catheter study using MIMIC-III
For example, the MIMIC team provides code for the MIMIC indwelling arterial catheter (IAC) aline study. The study uses the MIMIC-III dataset to reproduce findings from the published study, The Association Between Indwelling Arterial Catheters and Mortality in Hemodynamically Stable Patients With Respiratory Failure.
You can obtain the aline study code, as well as many other analytic examples, in the MIT Laboratory of Computational Physiology MIMIC Code Repository in GitHub. Learn more about the provided code in the JAMA paper, The MIMIC Code Repository: enabling reproducibility in critical care research.
The aline study code comprises about 1400 lines of SQL. It was developed for a PostgreSQL database, and is executed and further analyzed by Python and R code. You can run the aline study against the Parquet MIMIC-III dataset using Athena instead of PostgreSQL to answer the following questions:
- What modifications would be required, if any, to run the study using Athena instead of PostgreSQL?
- How would performance differ?
- How would cost differ?
Some SQL features used in the study work differently in Athena than they do in PostgreSQL. As a result, about 5% of the SQL statements needed modification. Specifically, the SQL statements require the following types of modification for use with Athena:
- Instead of using materialized views, in Athena, create a new table. For instance, the
CREATE MATERIALIZED VIEWstatement can be written asCREATE TABLE. - Instead of using schema context statements like
SET SEARCH_PATH, in Athena, explicitly declare the schema of referenced tables. For instance, instead of having aSET SEARCH_PATHstatement before your query and referencing tables without a schema as follows:
SET SEARCH_PATH TO mimiciii;
left join chartevents ce
You declare the schema as a part of every table reference:
left join mimiciii.chartevents ce
- Epoch time, as well as timestamp arithmetic, works differently in Athena than PostgreSQL. So, instead of arithmetic, use the date_diff() and specify seconds as the return value.
extract(epoch from endtime-starttime)/24.0/60.0/60.0
The preceding statement can be written as:
date_diff('second',starttime, endtime)/24.0/60.0/60.0
- Athena uses the “double” data type instead of the “numeric” data type.
ROUND( cast(f.height_first as numeric), 2) AS height_first,
The preceding statement can be written as:
ROUND( cast(f.height_first as double), 2) AS height_first,
- If VARCHAR fields are empty, they are not detected as NULL. Instead, compare a VARCHAR to an empty string. For instance, consider the following statement:
where ce.value IS NOT NULL
If ce.value is a VARCHAR, it can be written as the following:
where ce.value <> ''
After making these minor edits to the aline study SQL statements, the study executes successfully using Athena instead of PostgreSQL. The output produced from both methods is identical.
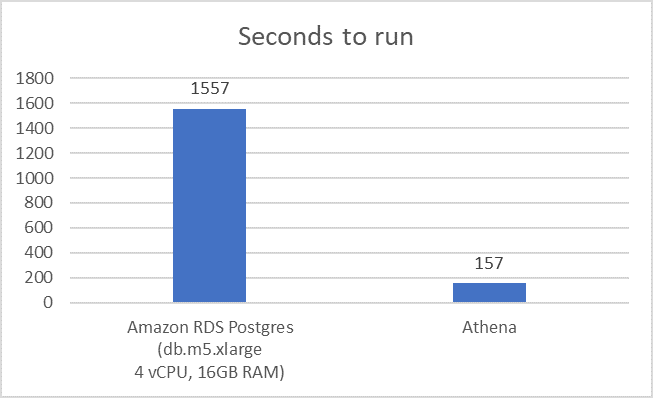
Next, consider the speed of Athena compared to PostgreSQL in analyzing the aline study. As shown in the following graph, the aline study using Athena queries of the Parquet-formatted MIMIC-III dataset run 10 times faster than queries of the same data in an Amazon RDS PostgreSQL database.
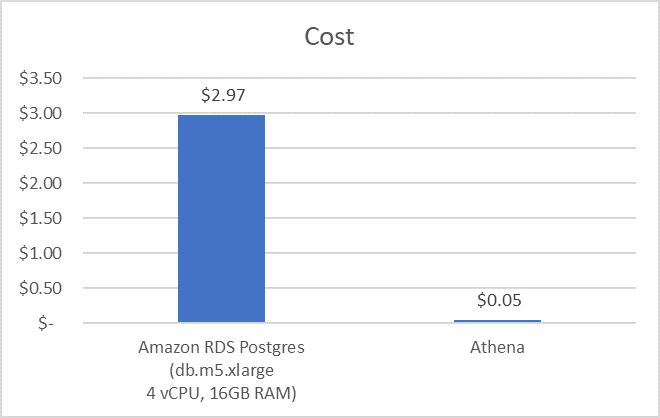
Compare the costs of running the aline study in Athena to running it in PostgreSQL. RDS PostgreSQL databases bill in one-second increments for the time the database runs, while Athena queries bill per gigabyte of data each query scans. Because of this fundamental pricing difference, you must make some assumptions to compare costs.
You could assume that running the aline study in RDS PostgreSQL involves the following steps, taking up an eight-hour working day:
- Deploy a new RDS PostgreSQL database.
- Load the MIMIC-III dataset.
- Connect a Jupyter notebook.
- Run the aline study.
- Terminate the RDS database.
You’d then compare the cost of running an RDS PostgreSQL database for eight hours (at $0.37 per hour, db.m5.xlarge with 100-GB EBS storage) to the cost of running the aline study against a dataset using Athena (9.93 GB of data scanned at $0.005/GB). As the following graph shows, this results in an almost 60x cost savings.
Working with MIMIC-III in AWS
MIMIC-III is a publicly available dataset containing detailed information about the clinical care of patients. For this reason, the MIMIC team requires that you complete a training course and submit a formal request before gaining access. For more information, see Requesting access.
After you obtain access to the MIMIC-III dataset, log in to the PhysioNet website and, under your profile settings, provide your AWS account ID. Then click the request access link, under Files, on the MIMIC-III Clinical Database page. You then have access to the MIMIC-III dataset in AWS.

Choose Launch Stack below to begin working with the MIMIC-III dataset in your AWS account. This launches an AWS CloudFormation template, which deploys an index of the MIMIC-III Parquet-formatted dataset into your AWS Glue Data Catalog. It also deploys an Amazon SageMaker notebook instance pre-loaded with the aline study code and many other MIMIC-provided analytic examples.
After the AWS CloudFormation template deploys, choose Outputs, and follow the link to access Jupyter Notebooks. From there, you can open and execute the aline study code by browsing the filesystem to the path ./mimic-code/notebooks/aline-aws/aline-awsathena.ipynb.
After you’ve completed your analysis, you can stop your Jupyter Notebook instance to suspend charges for compute. Any time that you want to continue working, just start the instance and continue where you left off.
Conclusion
Using the AWS Public Dataset program, MIT’s LCP provides quick and inexpensive access to the global biomedical research resources of the MIMIC-III dataset. Cloud-native analytic tools like AWS Glue and Athena accelerate research, and groups like the MIT Laboratory of Computational Physiology are pioneering data availability in modern data formats like Apache Parquet.
The analytic approaches and code demonstrated in this post can be applied generally to help increase agility and decrease cost. Consider using them as you enhance existing or perform new biomedical informatics research.
The content and opinions in this post are those of the third-party author and AWS is not responsible for the content or accuracy of this post.
About the Author
 James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.
James Wiggins is a senior healthcare solutions architect at AWS. He is passionate about using technology to help organizations positively impact world health. He also loves spending time with his wife and three children.
 Alistair Johnson is a research scientist at the Massachusetts Institute of Technology. Alistair has extensive experience and expertise in working with ICU data, having published the MIMIC-III and eICU-CRD datasets. His current research focuses on clinical epidemiology and machine learning for decision support.
Alistair Johnson is a research scientist at the Massachusetts Institute of Technology. Alistair has extensive experience and expertise in working with ICU data, having published the MIMIC-III and eICU-CRD datasets. His current research focuses on clinical epidemiology and machine learning for decision support.
