AWS for Industries
Pfizer’s Echocardiography Analysis Framework Reduces Time by +92% Using AWS
Blog is guest authored by Chong Duan, Amey Kelekar, and Mary Kate Montgomery from Pfizer
Echocardiography (echo) is an ultrasound-based, non-invasive imaging modality. Manual analysis of echo data is time consuming and susceptible to inter-reader variability. Pfizer has developed a fully automated analysis framework that significantly reduced analysis time by more than 92% using Amazon Web Services (AWS).
Echocardiography is widely used to assess cardiac structure and function in both clinical and preclinical research. In preclinical drug research and development, it is routinely used to assess the cardiac disease burden in animal models for heart failure and assess the cardiovascular safety of new drugs. The current workflow for preclinical echo analysis requires manual tracing of the endomyocardial border at end-systole and end-diastole frames.
Deep Learning Model and Results
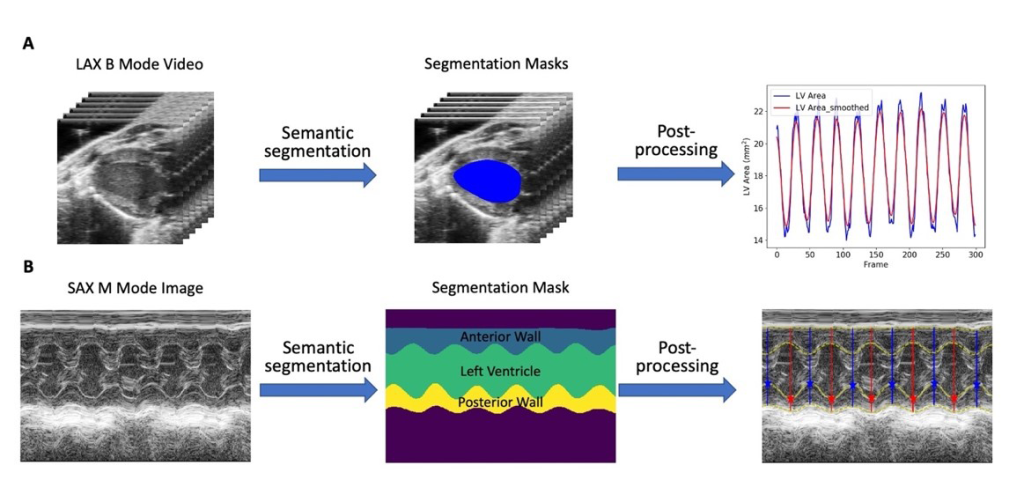 Figure 1 – Fully automated echocardiography analysis pipeline (Adapted from Fully Automated Mouse Echocardiography Analysis Using Deep Convolutional Neural Networks)
Figure 1 – Fully automated echocardiography analysis pipeline (Adapted from Fully Automated Mouse Echocardiography Analysis Using Deep Convolutional Neural Networks)
The Digital Science & Translational Imaging (DSTI) group and the Comparative Medicine (CM) group at Pfizer developed and validated a deep learning-based, fully automated, end-to-end echo analysis framework for B-mode and M-mode echo images/videos (Figure 1). Specifically, fully convolutional neural networks (adapted from U-net: Convolutional networks for biomedical image segmentation) were employed to segment the left ventricle and myocardial wall on either B-mode or M-mode images/videos. The left ventricle (LV) segmentation masks were then processed subsequently to obtain LV structure and function metrics including ejection fraction, fractional shortening, LV mass and volumes at different cardiac phases. These metrics, as well as an image of the segmentation results for quality control purposes, were generated for each B-mode or M-mode image/video and provided for researchers for further analysis.
This framework achieved excellent correlations (Pearson’s r = 0.85 – 0.99) when compared to the results from manual analysis. Further inter-reader variability analysis showed that this framework has better agreements with an expert analyst than both a trained analyst and a novice. The use of this tool reduced echo image analysis time by more than 92%.
Implementation
The analysis framework was built using AWS with scalability, flexibility, and reproducibility in mind.
Scalability
Echocardiography data are stored in Digital Imaging and Communications in Medicine (DICOM) format. The number of files can range from dozens to thousands. The team leveraged Amazon Simple Storage Service (Amazon S3), an object-based storage to store files of varying size. Amazon S3 has no limits on the number of files, which provided the flexibility they needed for the different file sizes.
To process the files AWS Batch, a fully managed batch processing service, was used. AWS Batch dynamically provisions the optimal Graphical Processing Unit (GPU) instance based on the volume and the requirements of the job created. This could support scaling to a larger GPU instance size if more files were present.
Flexibility
The process needed to be flexible to support changes to the application and services. Creating a continuous integration/continuous delivery (CI/CD) pipeline enabled the team to make changes to the application, test those changes, and deploy them in an automated way. Deploying the application in containers gave the team flexibility to define the specific requirements needed for their application.
Reproducibility
The echo analysis pipeline needed to be immutable to ensure consistent results. The team used AWS CloudFormation (CloudFormation), an infrastructure as code tool, to templatize AWS services to model their resources. CloudFormation handled provisioning and configuring the resources so they could focus on the analysis. CloudFormation templates could also be reused to build other applications.
The Analysis Framework
 Figure 2 – Echo analysis AWS deployment architecture
Figure 2 – Echo analysis AWS deployment architecture
The analysis starts when a user uploads echo data (DICOM files) into an input directory on a mounted file share through AWS Storage Gateway. AWS Storage Gateway is a hybrid cloud storage that allows easy integration with AWS storage services. The uploaded data is synced to Amazon S3. A daily cron job is triggered through Amazon EventBridge. Amazon EventBridge is a serverless, fully managed, and scalable event bus that allows applications to communicate using events. Amazon EventBridge uses a rule to execute an AWS Step Function workflow. AWS Step Functions is a serverless orchestration service that lets you combine AWS services to build business critical applications. AWS Step Functions breaks down workflows into steps to uses AWS Lambda. AWS Lambda is a serverless, event driven compute service that can scale to match demand allowing the team to handle processing of files without worrying about the underlying infrastructure.
The Step Functions workflow is as follows:
- AWS Batch will start and initialize a GPU instance, deploy, and install all the requirements, execute any scripts, and terminate the instance when the process is complete.
- The outputs (for example, cardiac parameter values and segmentation images) from the process are saved in an output directory in the S3 bucket. Processed DICOM files are also moved to an archive folder in an S3 bucket.
- A notification is sent out using Amazon Simple Notification Service (Amazon SNS). Amazon SNS will notify the team on whether the Step Function was executed successfully or not.
 Figure 3 – Continuous Integration/Deployment Architecture
Figure 3 – Continuous Integration/Deployment Architecture
Continuous Integration/Continuous Delivery (CI/CD)
To support any changes that need to be made to the analysis framework, the team also developed a robust CI/CD pipeline (Figure 3):
- When a developer makes changes to the code base, they push those changes to a GitHub repository.
- Changes pushed to GitHub will trigger AWS CodePipeline (CodePipeline), a fully managed continuous delivery service.
- CodePipeline compiles the code using AWS CodeBuild, a fully managed continuous integration service that runs tests and products software packages.
- When CodePipeline is done a container image is created and stored in Amazon Elastic Container Registry (Amazon ECR), a fully managed container registry.
- AWS Batch can then pull the most updated container image for the analysis pipeline.
Conclusion
Using AWS Services, the team was able to develop and deploy a fully automated echocardiography analysis tool that generates cardiac structural and function parameters consistent with manual analysis. This tool reduced analysis time by more than 92% and is currently being used by scientists at different research units at Pfizer to support drug research and development programs across therapeutic areas. With the flexibility and reproducibility that the deployment framework provides, it can be quickly adapted to other similar image analysis pipelines.
Further Reading
- Deploy digital biomarkers at scale in clinical trials using serverless services on AWS
- Orchestrating high performance computing with AWS Step Functions and AWS Batch
- Visit the AWS Health web page
 Chong Duan is an Associate Director, AI/ML Lead in the Digital Sciences & Translational Imaging (DSTI) group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. Chong holds a B.S. degree in Chemistry from Nankai University, a M.S. degree in Computer Science from Georgia Institute of Technology, and a Ph.D. degree in MRI Physics from Washington University in St. Louis. He currently leads the development and deployment of novel machine learning methods for biomedical image analysis in drug R&D at Pfizer.
Chong Duan is an Associate Director, AI/ML Lead in the Digital Sciences & Translational Imaging (DSTI) group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. Chong holds a B.S. degree in Chemistry from Nankai University, a M.S. degree in Computer Science from Georgia Institute of Technology, and a Ph.D. degree in MRI Physics from Washington University in St. Louis. He currently leads the development and deployment of novel machine learning methods for biomedical image analysis in drug R&D at Pfizer.
 Amey Kelekar is a Manager, Medical Informatics in the Digital Sciences & Translational Imaging (DSTI) group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. His core focus includes Data management and cloud operations activities for DSTI’s clinical and non-clinical trials. Amey holds a Bachelor of Engineering degree in Computer Engineering from Mumbai University and a Master of Science degree in Information Systems from Northeastern University.
Amey Kelekar is a Manager, Medical Informatics in the Digital Sciences & Translational Imaging (DSTI) group within Early Clinical Development in Worldwide Research, Development, and Medical at Pfizer. His core focus includes Data management and cloud operations activities for DSTI’s clinical and non-clinical trials. Amey holds a Bachelor of Engineering degree in Computer Engineering from Mumbai University and a Master of Science degree in Information Systems from Northeastern University.
 Mary Kate Montgomery is an Image Analysis Senior Scientist in the Global Science and Technology group within Comparative Medicine in Worldwide Research, Development, and Medical at Pfizer. Her work mainly focuses on automating data analysis processes for pre-clinical imaging studies. Mary Kate holds a Bachelor of Science in Biomedical Engineering from Columbia University and a Master of Advanced Studies in Data Science and Engineering from the University of California San Diego.
Mary Kate Montgomery is an Image Analysis Senior Scientist in the Global Science and Technology group within Comparative Medicine in Worldwide Research, Development, and Medical at Pfizer. Her work mainly focuses on automating data analysis processes for pre-clinical imaging studies. Mary Kate holds a Bachelor of Science in Biomedical Engineering from Columbia University and a Master of Advanced Studies in Data Science and Engineering from the University of California San Diego.