AWS Machine Learning Blog
Running on-demand, serverless Apache Spark data processing jobs using Amazon SageMaker managed Spark containers and the Amazon SageMaker SDK
July 2023: This post was reviewed for accuracy.
Apache Spark is a unified analytics engine for large scale, distributed data processing. Typically, businesses with Spark-based workloads on AWS use their own stack built on top of Amazon Elastic Compute Cloud (Amazon EC2), or Amazon EMR to run and scale Apache Spark, Hive, Presto, and other big data frameworks. This is useful for persistent workloads, in which you want these Spark clusters to be up and running 24/7, or at best, would have to come up with an architecture to spin up and spin down the cluster on a schedule or on demand.
Amazon SageMaker Processing lets you easily run preprocessing, postprocessing, model evaluation or other fairly generic transform workloads on a fully managed infrastructure. Previously, Amazon SageMaker Processing included a built-in container for Scikit-learn style preprocessing. For using other libraries like Spark, you have the flexibility to bring in your own Docker containers. Amazon SageMaker Processing jobs can also be part of your Step Functions workflow for ML involving pre- and post-processing steps. For more information, see AWS Step Functions adds support for Amazon SageMaker Processing.
Several machine learning(ML) workflows involve preprocessing data with Spark (or other libraries) and then passing in training data to a training step. The following workflow shows an Extract, Transform and Load (ETL) step that leads to model training and finally to model endpoint deployment using AWS Step Functions.

Including Spark steps in such workflows requires additional steps to provision and set up these clusters. Alternatively, you can do this using AWS Glue, a fully managed ETL service that makes it easy for customers to write Python or Scala based Spark scripts to preprocess data for ML training.
We’re happy to add a managed Spark container and associated SDK enhancements to Amazon SageMaker Processing, which lets you perform large scale, distributed processing on Spark by simply submitting a PySpark or Java/Scala Spark application. You can use this feature in Amazon SageMaker Studio and Amazon SageMaker notebook instances.
To demonstrate, the following code example runs a PySpark script on Amazon SageMaker Processing by using the PySparkProcessor:
We can look at this example in some more detail. The PySpark script name ‘preprocess.py’ such as the one shown below, that loads a large CSV file from Amazon Simple Storage Service (Amazon S3) into a Spark dataframe, fits and transforms this dataframe into an output dataframe, and converts and saves a CSV back to Amazon S3:
You can easily start a Spark based processing job by using the PySparkProcessor() class as shown below:
When running this in Amazon SageMaker Studio or Amazon SageMaker notebook instance, the output shows the job’s progress:
In Amazon SageMaker Studio, you can describe your processing jobs and view relevant details by choosing the processing job name (right-click), and choosing Open in trial details.
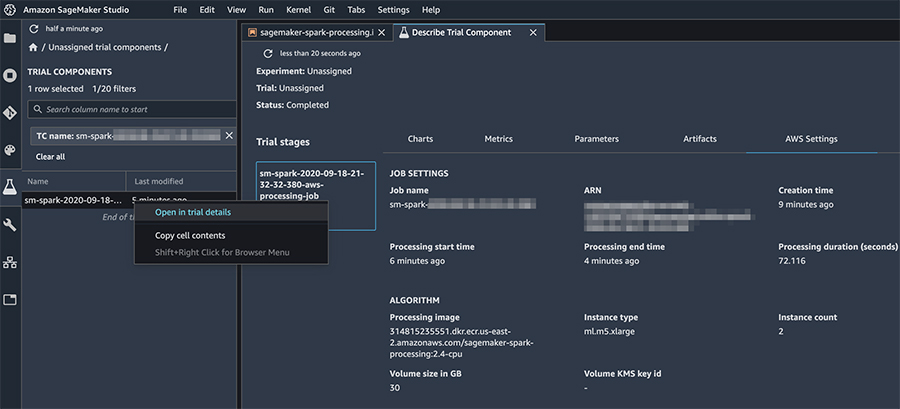
You can also track the processing job’s settings, logs, and metrics on the Amazon SageMaker console as shown in the following screenshot.

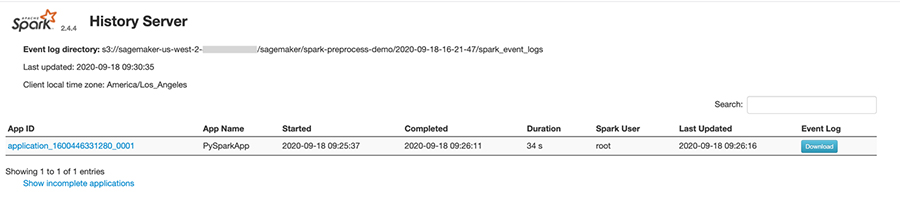
After a job completes, if the spark_event_logs_s3_uri was specified in the run() function, the Spark UI can be viewed by running the history server:
spark_processor.start_history_server()
If run from an Amazon SageMaker Notebook instance, the output will include a proxy URL where the history server can be accessed:
Visiting this URL will bring you to the history server web interface as shown in the screenshot below:
Additional python and jar file dependencies can also be specified in your Spark jobs. For example, if you want to serialize an MLeap model, you can specify these additional dependencies by modifying the call to the run() function of PySparkProcessor:
Finally, overriding Spark configuration is crucial for several tasks such as tuning your Spark application or configuring the Hive metastore. You can override Spark, Hive, Hadoop configurations using our Python SDK.
For example, the following code overrides spark.executor.memory and spark.executor.cores:
Try out this example on your own by navigating to the examples tab in your Amazon SageMaker notebook instance, or by cloning the Amazon SageMaker examples directory and navigating to the folder with Amazon SageMaker Processing examples.
Additionally, you can set up an end-to-end Spark workflow for your use cases using Amazon SageMaker and other AWS services:
- Connecting your notebook instances using Livy to an EMR Spark cluster – https://aws.amazon.com/blogs/machine-learning/build-amazon-sagemaker-notebooks-backed-by-spark-in-amazon-emr/
- Deploying MLeap serialized models on SageMaker endpoints using SparkML serving – https://sagemaker.readthedocs.io/en/stable/frameworks/sparkml/sagemaker.sparkml.html
- Using Spark on SageMaker Processing as shown in this post for Spark based preprocessing, training and post processing jobs – https://github.com/awslabs/amazon-sagemaker-examples/blob/master/sagemaker_processing/spark_distributed_data_processing/sagemaker-spark-processing.ipynb
- Using your Spark based Processing jobs in a Step functions workflow – https://docs.aws.amazon.com/step-functions/latest/dg/concepts-python-sdk.html and https://docs.aws.amazon.com/step-functions/latest/dg/connect-sagemaker.html
Conclusion
Amazon SageMaker makes extensive use of Docker containers to allow users to build a runtime environment for data preparation, training, and inference code. Amazon SageMaker’s built-in Spark container for Amazon SageMaker Processing provides a managed Spark runtime including all library components and dependencies needed to run distributed data processing workloads. The example discussed in the blog shows how developers and data scientists can take advantage of the built-in Spark container on Amazon SageMaker to focus on more important aspects of preparing and preprocessing data. Instead of spending time tuning, scaling, or managing Spark infrastructure, developers can focus on core implementation.
About the Authors
 Shreyas Subramanian is a AI/ML specialist Solutions Architect, and helps customers by using Machine Learning to solve their business challenges using the AWS platform.
Shreyas Subramanian is a AI/ML specialist Solutions Architect, and helps customers by using Machine Learning to solve their business challenges using the AWS platform.
 Andrew Packer is a Software Engineer in Amazon AI where he is excited about building scalable, distributed machine learning infrastructure for the masses. In his spare time, he likes playing guitar and exploring the PNW.
Andrew Packer is a Software Engineer in Amazon AI where he is excited about building scalable, distributed machine learning infrastructure for the masses. In his spare time, he likes playing guitar and exploring the PNW.
 Vidhi Kastuar is a Sr. Product Manager for Amazon SageMaker, focusing on making machine learning and artificial intelligence simple, easy to use and scalable for all users and businesses. Prior to AWS, Vidhi was Director of Product Management at Veritas Technologies. For fun outside work, Vidhi loves to sketch and paint, work as a career coach, and spend time with her family and friends.
Vidhi Kastuar is a Sr. Product Manager for Amazon SageMaker, focusing on making machine learning and artificial intelligence simple, easy to use and scalable for all users and businesses. Prior to AWS, Vidhi was Director of Product Management at Veritas Technologies. For fun outside work, Vidhi loves to sketch and paint, work as a career coach, and spend time with her family and friends.