AWS Machine Learning Blog
Detect mitotic figures in whole slide images with Amazon Rekognition
Even after more than a hundred years after its introduction, histology remains the gold standard in tumor diagnosis and prognosis. Anatomic pathologists evaluate histology to stratify cancer patients into different groups depending on their tumor genotypes and phenotypes, and their clinical outcome [1,2]. However, human evaluation of histological slides is subjective and not repeatable [3]. Furthermore, histological assessment is a time-consuming process that requires highly trained professionals.
With significant technological advances in the last decade, techniques such as whole slide imaging (WSI) and deep learning (DL) are now widely available. WSI is the scanning of conventional microscopy glass slides to produce a single, high-resolution image from those slides. This allows for the digitization and collection of large sets of pathology images, which would have been prohibitively time-consuming and expensive. The availability of such datasets creates new and innovative ways of accelerating diagnosis by using techniques such as machine learning (ML) to aid pathologists to accelerate diagnoses by quickly identifying features of interest.
In this post, we will explore how developers without previous ML experience can use Amazon Rekognition Custom Labels to train a model that classifies cellular features. Amazon Rekognition Custom Labels is a feature of Amazon Rekognition that enables you to build your own specialized ML-based image analysis capabilities to detect unique objects and scenes integral to your specific use case. In particular, we use a dataset containing whole slide images of canine mammary carcinoma [1] to demonstrate how to process these images and train a model that detects mitotic figures. This dataset has been used with permission from Prof. Dr. Marc Aubreville, who has kindly agreed to allow us to use it for this post. For more information, see the Acknowledgments section at the end of this post.
Solution Overview
The solution consists of two components:
- An Amazon Rekognition Custom Labels model — To enable Amazon Rekognition to detect mitotic figures, we complete the following steps:
- Sample the WSI dataset to produce adequately sized images using Amazon SageMaker Studio and a Python code running on a Jupyter notebook. Studio is a web-based, integrated development environment (IDE) for ML that provides all the tools you need to take your models from experimentation to production while boosting your productivity. We will use Studio to split the images into smaller ones to train our model.
- Train a Amazon Rekognition Custom Labels model to recognize mitotic figures in hematoxylin-eosin samples using the data prepared in the previous step.
- A frontend application — To demonstrate how to use a model like the one we trained in the previous step, we complete the following steps:
- Write a small sample web application using Python and Streamlit.
- Deploy it as a container using AWS Fargate and Amazon Elastic Container Service (Amazon ECS).
- Build a continuous integration, continuous delivery (CI/CD) pipeline to automate the deployment of the application using AWS CodeBuild and AWS CodePipeline.
- Write the entire deployment script as an AWS Cloud Development Kit (AWS CDK) script.
The following diagram illustrates the solution architecture.
All the necessary resources to deploy the implementation discussed in this post and the code for the whole section are available on GitHub. You can clone or fork the repository, make any changes you desire, and run it yourself.
In the next steps, we walk through the code to understand the different steps involved in obtaining and preparing the data, training the model, and using it from a sample application.
Costs
When running the steps in this walkthrough, you incur small costs from using the following AWS services:
- Amazon Rekognition
- AWS Fargate
- Application Load Balancer
- AWS Secrets Manager
Additionally, if no longer within the Free Tier period or conditions, you may incur costs from the following services:
- CodePipeline
- CodeBuild
- Amazon ECR
- Amazon SageMaker
If you complete the cleanup steps correctly after finishing this walkthrough, you may expect costs to be less than 10 USD, if the Amazon Rekognition Custom Labels model and the web application run for one hour or less.
Prerequisites
To complete all steps, you need the following:
- An AWS account
- The AWS Command Line Interface (AWS CLI) installed and configured to interact with AWS services locally
- The AWS CDK installed and configured to deploy resources to your AWS account
Training the mitotic figure classification model
We run all the steps required to train the model from a Studio notebook. If you have never used Studio before, you may need to onboard first. For more information, see Onboard Quickly to Amazon SageMaker Studio.
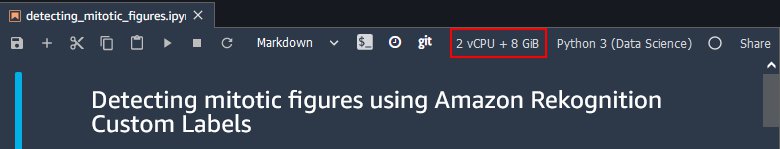
Some of the following steps require more RAM than what is available in a standard ml.t3.medium notebook. Make sure that you have selected an ml.m5.large notebook. You should see a 2 vCPU + 8 GiB indication on the upper right corner of the page.
The code for this section is available as a Jupyter notebook file.
After onboarding to Studio, follow these instructions to grant Studio the necessary permissions to call Amazon Rekognition on your behalf.
Dependencies
To begin with, we need to complete the following steps:
- Update Linux packages and install the required dependencies, such as OpenSlide:
- Install the fastai and SlideRunner libraries using pip:
- Download the dataset (we provide a script to do this automatically):
Process the dataset
We will begin by importing some of the packages that we use throughout the data preparation stage. Then, we download and load the annotation database for this dataset. This database contains the positions in the whole slide images of the mitotic figures (the features we want to classify). See the following code:
Because we’re using SageMaker, we create a new SageMaker session object to ease tasks such as uploading our dataset to an Amazon Simple Storage Service (Amazon S3) bucket. We also use the S3 bucket that SageMaker creates by default to upload our processed image files.
The slidelist_test array contains the IDs of the slides that we use as part of the test dataset to evaluate the performance of the trained model. See the following code:
The next step is to obtain a set of areas of training and test slides, along with the labels in them, from which we can take smaller areas to use to train our model. The code for get_slides is in the sampling.py file in GitHub.
We want to randomly sample from the training and test slides. We use the lists of training and test slides and randomly select n_training_images times a file for training, and n_test_images times a file for test:
Next, we create a directory for training images and one for test images:
Before we produce the smaller images needed to train the model, we need some helper code that produces the metadata needed to describe the training and test data. The following code makes sure that a given bounding box surrounding the features of interest (mitotic figures) are well within the zone we’re cutting, and produces a line of JSON that describes the image and the features in it in Amazon SageMaker Ground Truth format, which is the format Amazon Rekognition Custom Labels requires. For more information about this manifest file for object detection, see Object localization in manifest files.
With the generate_annotations function in place, we can write the code to produce the training and test images:
The last step towards having all of the required data is to write a manifest.json file for each of the datasets:
Transfer the files to S3
We use the upload_data method that the SageMaker session object exposes to upload the images and manifest files to the default SageMaker S3 bucket:
Train an Amazon Rekognition Custom Labels model
With the data already in Amazon S3, we can get to training a custom model. We use the Boto3 library to create an Amazon Rekognition client and create a project:
With the project ready to use, now you need a project version that points to the training and test datasets in Amazon S3. Each version ideally points to different datasets (or different versions of it). This enables us to have different versions of a model, compare their performance, and switch between them as needed. See the following code:
After we create the project version, Amazon Rekognition automatically starts the training process. The training time depends on several features, such as the size of the images and the number of them, the number of classes, and so on. In this case, for 500 images, the training takes about 90 minutes to finish.
Test the model
After training, every model in Amazon Rekognition Custom Labels is in the STOPPED state. To use it for inference, you need to start it. We get the project version ARN from the project version description and pass it over to the start_project_version. Notice the MinInferenceUnits parameter — we start with one inference unit. The actual maximum number of transactions per second (TPS) that this inference unit supports depends on the complexity of your model. To learn more about TPS, refer to this blog post.
When your project version is listed as RUNNING, you can start sending images to Amazon Rekognition for inference.
We use one of the files in the test dataset to test the newly started model. You can use any suitable PNG or JPEG file instead.
Streamlit application
To demonstrate the integration with Amazon Rekognition, we use a very simple Python application. We use the Streamlit library to build a spartan user interface, where we prompt the user to upload an image file.
We use the Boto3 library and the detect_custom_labels method, together with the project version ARN, to invoke the inference endpoint. The response is a JSON document that contains the positions and classes of the different objects detected in the image. In our case, these are the mitotic figures that the algorithm has found in the image we sent to the endpoint. See the following code:
Deploy the application to AWS
To deploy the application, we use an AWS CDK script. The whole project can be found on GitHub . Let’s look at the different resources deployed by the script.
Create an Amazon ECR repository
As the first step towards setting up our deployment, we create an Amazon ECR repository, where we can store our application container images:
Create and store your GitHub token in AWS Secrets Manager
CodePipeline needs a GitHub Personal Access Token to monitor your GitHub repository for changes and pull code. To create the token, follow the instructions in the GitHub documentation. The token requires the following GitHub scopes:
- The
reposcope, which is used for full control to read and pull artifacts from public and private repositories into a pipeline. - The
admin:repo_hookscope, which is used for full control of repository hooks.
After creating the token, store it in a new secret in AWS Secrets Manager as follows:
Write configuration parameters to AWS Systems Manager Parameter Store
The AWS CDK script reads some configuration parameters from AWS Systems Manager Parameter Store, such as the name and owner of the GitHub repository, and target account and Region. Before launching the AWS CDK script, you need to create these parameters in your own account.
You can do that by using the AWS CLI. Simply invoke the put-parameter command with a name, a value, and the type of the parameter:
The following is a list of all parameters required by the AWS CDK script. All of them are of type String:
- /rek_wsi/prod/accountId — The ID of the account where we deploy the application.
- /rek_wsi/prod/ecr_repo_name — The name of the Amazon ECR repository where the container images are stored.
- /rek_wsi/prod/github/branch — The branch in the GitHub repository from which CodePipeline needs to pull the code.
- /rek_wsi/prod/github/owner — The owner of the GitHub repository.
- /rek_wsi/prod/github/repo — The name of the GitHub repository where our code is stored.
- /rek_wsi/prod/github/token — The name or ARN of the secret in Secrets Manager that contains your GitHub authentication token. This is necessary for CodePipeline to be able to communicate with GitHub.
- /rek_wsi/prod/region — The region where we will deploy the application.
Notice the prod segment in all parameter names. Although we do not need this level of detail for such a simple example, it will enable to reuse this approach with other projects where different environments may be necessary.
Resources created by the AWS CDK script
We need our application, running in a Fargate task, to have permissions to invoke Amazon Rekognition. So we first create an AWS Identity and Access Management (IAM) Task Role with the RekognitionReadOnlyPolicy policy attached to it. Notice that the assumed_by parameter in the following code takes the ecs-tasks.amazonaws.com service principal. This is because we’re using Amazon ECS as the orchestrator, so we need Amazon ECS to assume the role and pass the credentials to the Fargate task.
Once built, our application container image sits in a private Amazon ECR repository. We need an object that describes it that we can pass when creating the Fargate service:
We create a new VPC and cluster for this application. You can modify this part to use your own VPC by using the from_lookup method of the Vpc class:
Now that we have a VPC and cluster to deploy to, we create the Fargate service. We use 0.25 vCPU and 512 MB RAM for this task, and we place a public Application Load Balancer (ALB) in front of it. Once deployed, we use the ALB CNAME to access the application. See the following code:
To automatically build and deploy a new container image every time we push code to our main branch, we create a simple pipeline consisting of a GitHub source action and a build step. Here is where we use the secrets we stored in AWS Secrets Manager and AWS Systems Manager Parameter Store in the previous steps.
CodeBuild needs permissions to push container images to Amazon ECR. To grant these permissions, we add the AmazonEC2ContainerRegistryFullAccess policy to a bespoke IAM role that the CodeBuild service principal can assume:
The CodeBuild project logs into the private Amazon ECR repository, builds the Docker image with the Streamlit application, and pushes the image into the repository together with an appspec.yaml and an imagedefinitions.json file.
The appspec.yaml file describes the task (port, Fargate platform version, and so on), while the imagedefinitions.json file maps the names of the container images to their corresponding Amazon ECR URI. See the following code:
Finally, we put the different pipeline stages together. The last action is the EcsDeployAction, which takes the container image built in the previous stage and does a rolling update of the tasks in our ECS cluster:
Cleanup
To avoid incurring future costs, clean up the resources you created as part of this solution.
Amazon Rekognition Custom Labels model
Before you shut down your Studio notebook, make sure you stop the Amazon Rekognition Custom Labels model. If you don’t, it continues to incur costs.
Alternatively, you can use the Amazon Rekognition console to stop the service:
- On the Amazon Rekognition console, choose Use Custom Labels in the navigation pane.
- Choose Projects in the navigation pane.
- Choose version 1 of the
rek-mitotic-figures-workshopproject. - On the Use Model tab, choose Stop.
Streamlit application
To destroy all resources associated to the Streamlit application, run the following code from the AWS CDK application directory:
AWS Secrets Manager
To delete the GitHub token, follow the instructions in the documentation.
Conclusion
In this post, we walked through the necessary steps to train a Amazon Rekognition Custom Labels model for a digital pathology application using real-world data. We then learned how to use the model from a simple application deployed from a CI/CD pipeline to Fargate.
Amazon Rekognition Custom Labels enables you to build ML-enabled healthcare applications that you can easily build and deploy using services like Fargate, CodeBuild, and CodePipeline.
Can you think of any applications to help researchers, doctors, or their patients to make their lives easier? If so, use the code in this walkthrough to build your next application. And if you have any questions, please share them in the comments section.
Acknowledgments
We would like to thank Prof. Dr. Marc Aubreville for kindly giving us permission to use the MITOS_WSI_CMC dataset for this blog post. The dataset can be found on GitHub.
References
[1] Aubreville, M., Bertram, C.A., Donovan, T.A. et al. A completely annotated whole slide image dataset of canine breast cancer to aid human breast cancer research. Sci Data 7, 417 (2020). https://doi.org/10.1038/s41597-020-00756-z
[2] Khened, M., Kori, A., Rajkumar, H. et al. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci Rep 11, 11579 (2021). https://doi.org/10.1038/s41598-021-90444-8
[3] PNAS March 27, 2018 115 (13) E2970-E2979; first published March 12, 2018; https://doi.org/10.1073/pnas.1717139115
About the Author
 Pablo Nuñez Pölcher, MSc, is a Senior Solutions Architect working for the Public Sector team with Amazon Web Services. Pablo focuses on helping healthcare public sector customers build new, innovative products on AWS in accordance with best practices. He received his M.Sc. in Biological Sciences from Universidad de Buenos Aires. In his spare time, he enjoys cycling and tinkering with ML-enabled embedded devices.
Pablo Nuñez Pölcher, MSc, is a Senior Solutions Architect working for the Public Sector team with Amazon Web Services. Pablo focuses on helping healthcare public sector customers build new, innovative products on AWS in accordance with best practices. He received his M.Sc. in Biological Sciences from Universidad de Buenos Aires. In his spare time, he enjoys cycling and tinkering with ML-enabled embedded devices.
 Razvan Ionasec, PhD, MBA, is the technical leader for healthcare at Amazon Web Services in Europe, Middle East, and Africa. His work focuses on helping healthcare customers solve business problems by leveraging technology. Previously, Razvan was the global head of artificial intelligence (AI) products at Siemens Healthineers in charge of AI-Rad Companion, the family of AI-powered and cloud-based digital health solutions for imaging. He holds 30+ patents in AI/ML for medical imaging and has published 70+ international peer-reviewed technical and clinical publications on computer vision, computational modeling, and medical image analysis. Razvan received his PhD in Computer Science from the Technical University Munich and MBA from University of Cambridge, Judge Business School.
Razvan Ionasec, PhD, MBA, is the technical leader for healthcare at Amazon Web Services in Europe, Middle East, and Africa. His work focuses on helping healthcare customers solve business problems by leveraging technology. Previously, Razvan was the global head of artificial intelligence (AI) products at Siemens Healthineers in charge of AI-Rad Companion, the family of AI-powered and cloud-based digital health solutions for imaging. He holds 30+ patents in AI/ML for medical imaging and has published 70+ international peer-reviewed technical and clinical publications on computer vision, computational modeling, and medical image analysis. Razvan received his PhD in Computer Science from the Technical University Munich and MBA from University of Cambridge, Judge Business School.